Directed evolution of enzymes and antibodies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Surface Expression of Anti-digoxin Single Chain Fv Antibodies
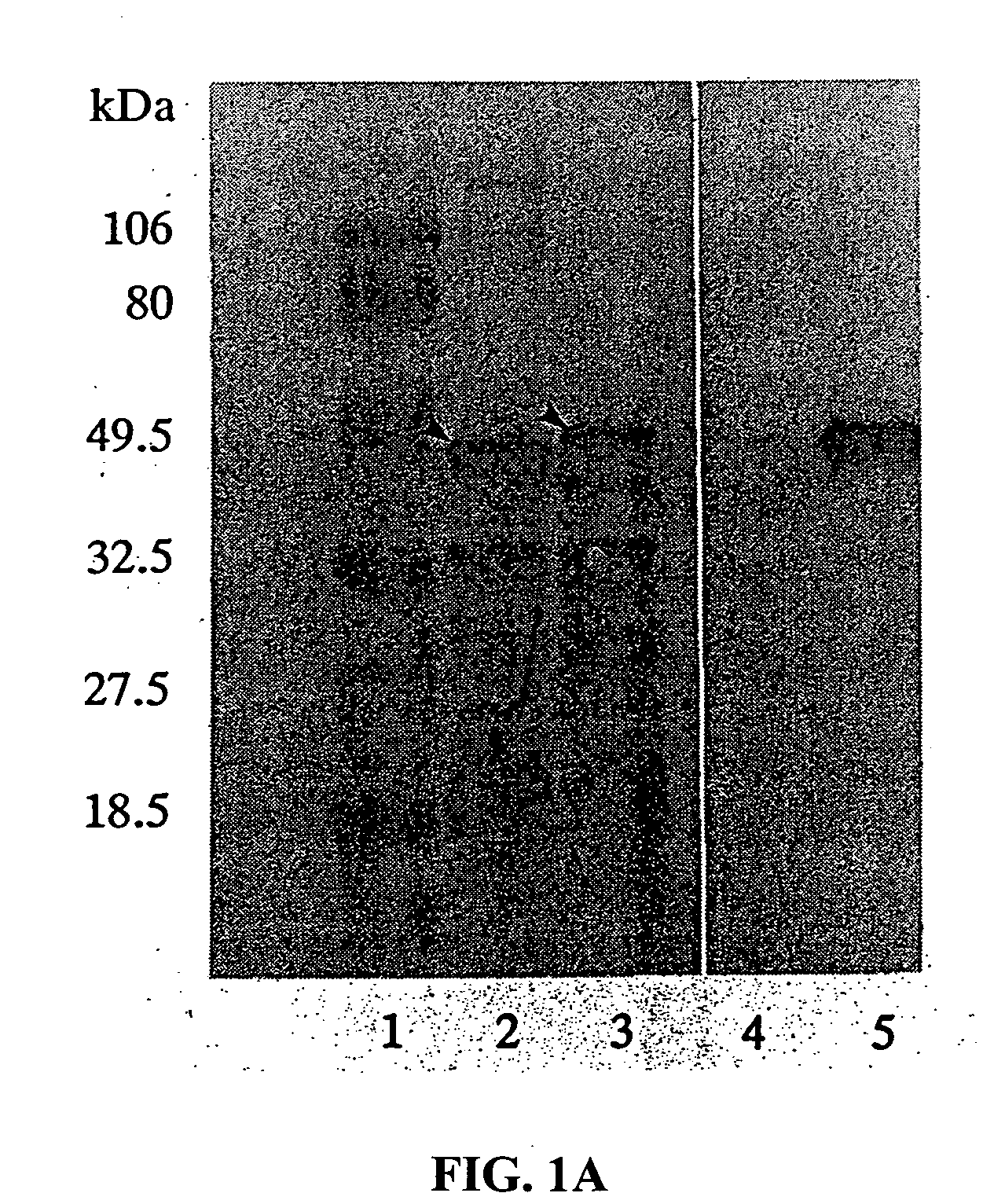
[0217]E. coli strain JM109 [endA1 recA1 gyrA thi-1 hsdR17 (rk−, Mk+) relA1 supE44 Δ(lac-proAB) / F′ traD36proAB laclq lacZΔM15] was used for all studies. pTX101 codes for an Lpp-OmpA-β-lactamase fusion (Francisco et al., 1992). pTX152 codes for an Lpp-OmpA-scFv(digoxin) fusion, where the scFv(digoxin) is an anti-digoxin single chain Fv consisting of the heavy- and light-chain variable regions (VH and VL). The VH and VL, joined by a 15 amino acid [(Gly)4Ser]3 linker (Huston et al., 1988), were amplified from messenger RNA isolated from two separate anti-digoxin hybridomas. An 11 amino acid peptide from the Herpes Simplex Virus glycoprotein (Novagen) was introduced at the C-terminus of the scFv for analytical purposes. The presence of the HSV peptide allowed detection of the scFv(digoxin) protein by reaction with a monoclonal antibody specific for the 11 amino acid epitope. The sequence of the single chain Fv antibody fragmen...
example 2
Enrichment of Cells Displaying scFv Antibodies by FACS
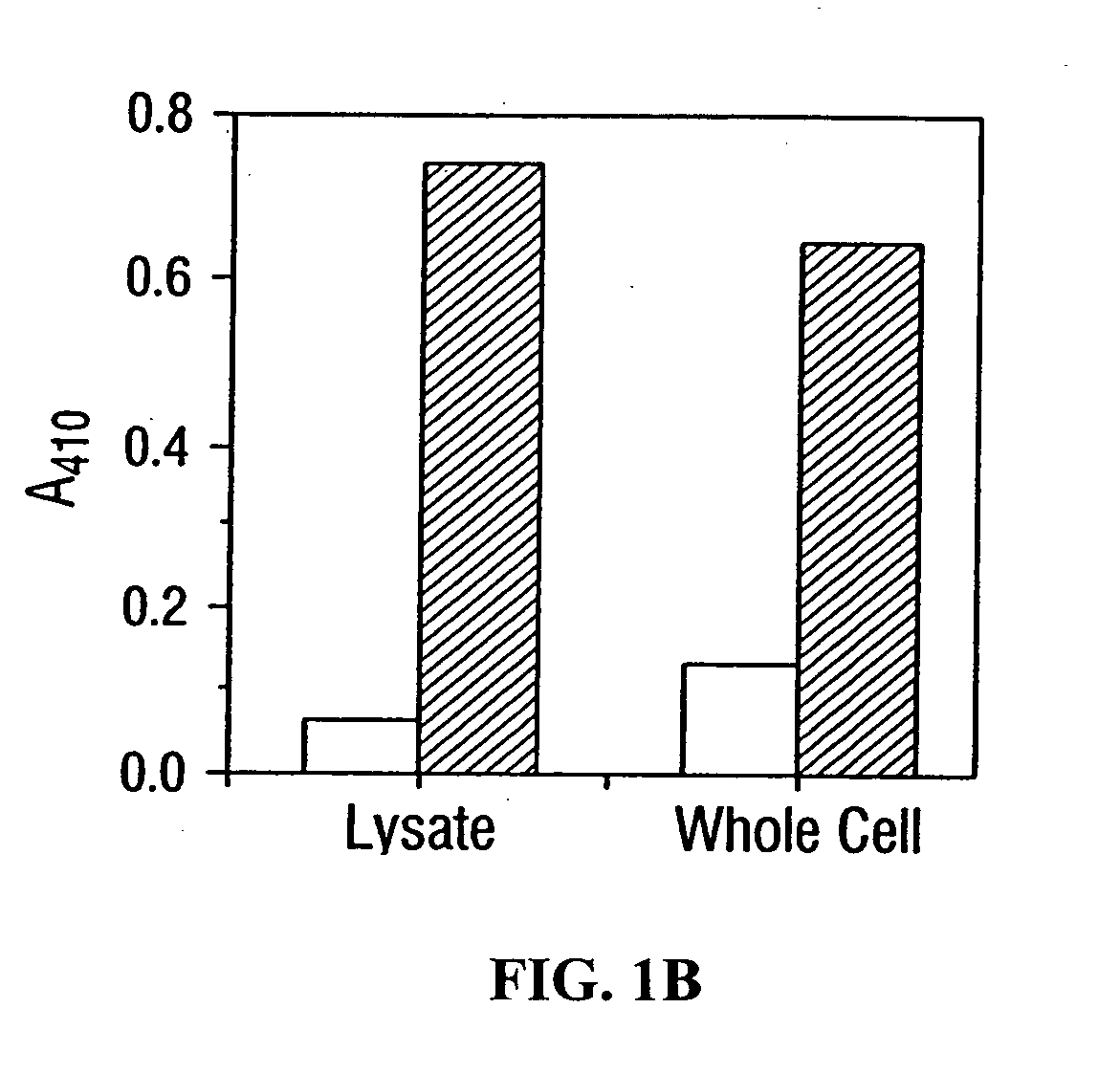
[0232] Antibody expressing cells were sorted essentially quantitatively from an excess of control E. Coli in a single step. Specifically, in mixtures containing JM109 / pTX101 control cells at an excess of either 100:1 or 1,000:1, the fraction of the total population that was sorted in the high fluorescence intensity window was 1.1% and 0.1% respectively (after subtracting the background), as expected from the ratio of input cells.
[0233] The use of FACS for isolating rare clones from a very large excess of background was also demonstrated. JM109 / pTX101 (cells displaying an unrelated protein on the cell surface) and JM101 / pTX152 (cells displaying the scFv(digoxin) antibodies) were mixed at a ratio of 100,000:1 and labeled with digoxin-FITC. Following washing to remove any non-specific binding of the digoxin-fluorescein conjugate on the control E. Coli, 500,000 cells were run through the FACSort flow cytometer. A wide sorting gate,...
example 3
Surface Expression of Anti-Digoxin Antibody Incorporating Protease Cleavage Site
[0238] A construct similar to that used for surface expression of scFv (digoxin) can be modified to incorporate a protease cleavage site. For example, the recognition sequence of enterokinase [(Asp)4-Ile-Arg] can be introduced in the Lpp-OmpA(46-159)-scFv between the OmpA(46-159) and the scFv domains. The protease cleavage site at the N-terminal of the scFv antibody domain of the fusion protein is then used to release the scFv antibody in soluble form following treatment of the cells with the appropriate proteolytic enzyme. Because the outer membrane of E. coli serves as a protective barrier to the action of externally added proteases, very few contaminating proteins will be present in the culture supernatant. A single colony expressing a desired single chain Fv antibody can be grown in liquid media and harvested by centrifugation after overnight growth at 24° C. The cells are resuspended in buffer to m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Size | aaaaa | aaaaa |
| Immunogenicity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com