Miniaturized lateral flow device for rapid and sensitive detection of proteins or nucleic acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Detection of DNA Hybridization Over Broad Range of Capture Oligonucleotide Deposition Concentrations on DNA Dipstick
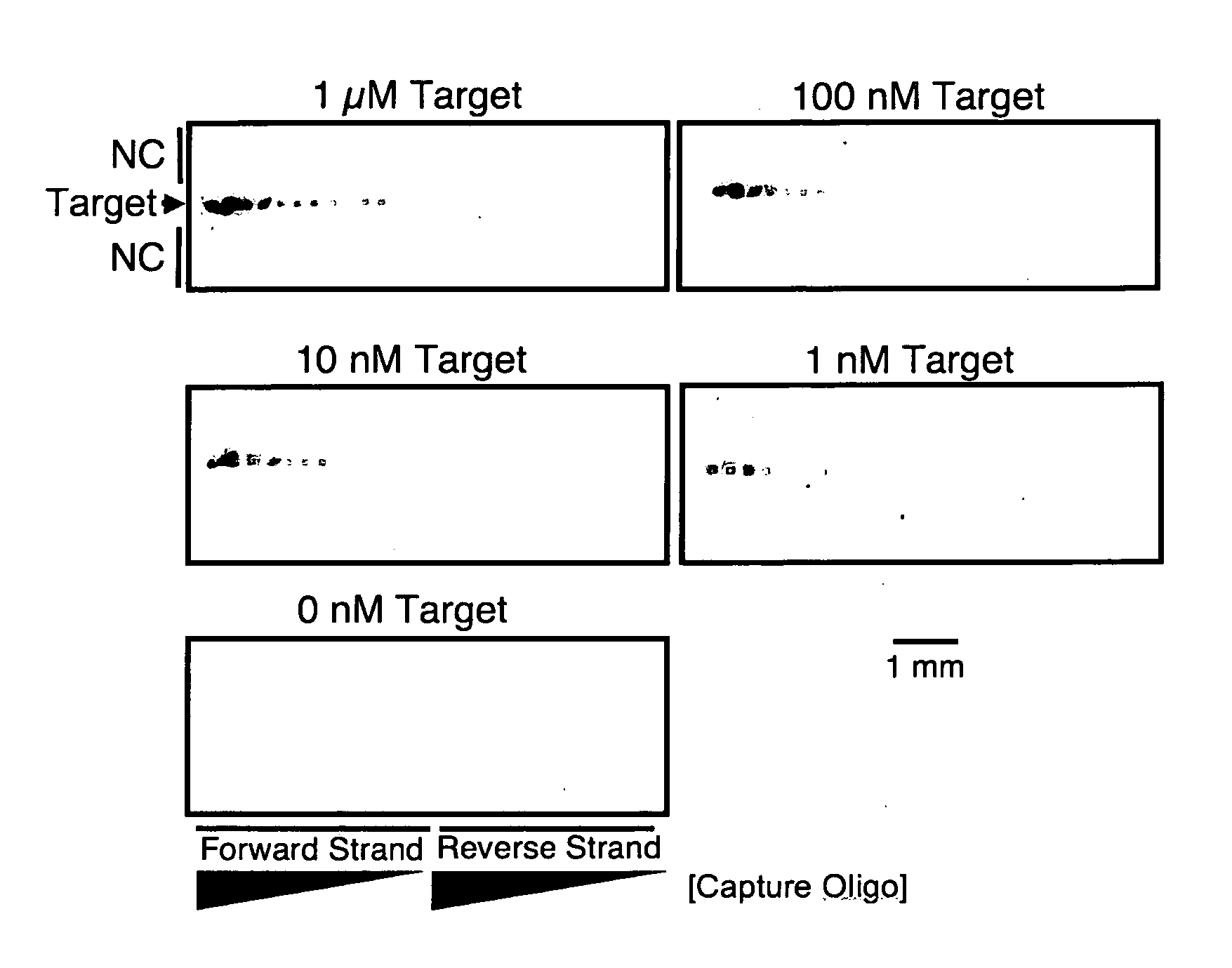
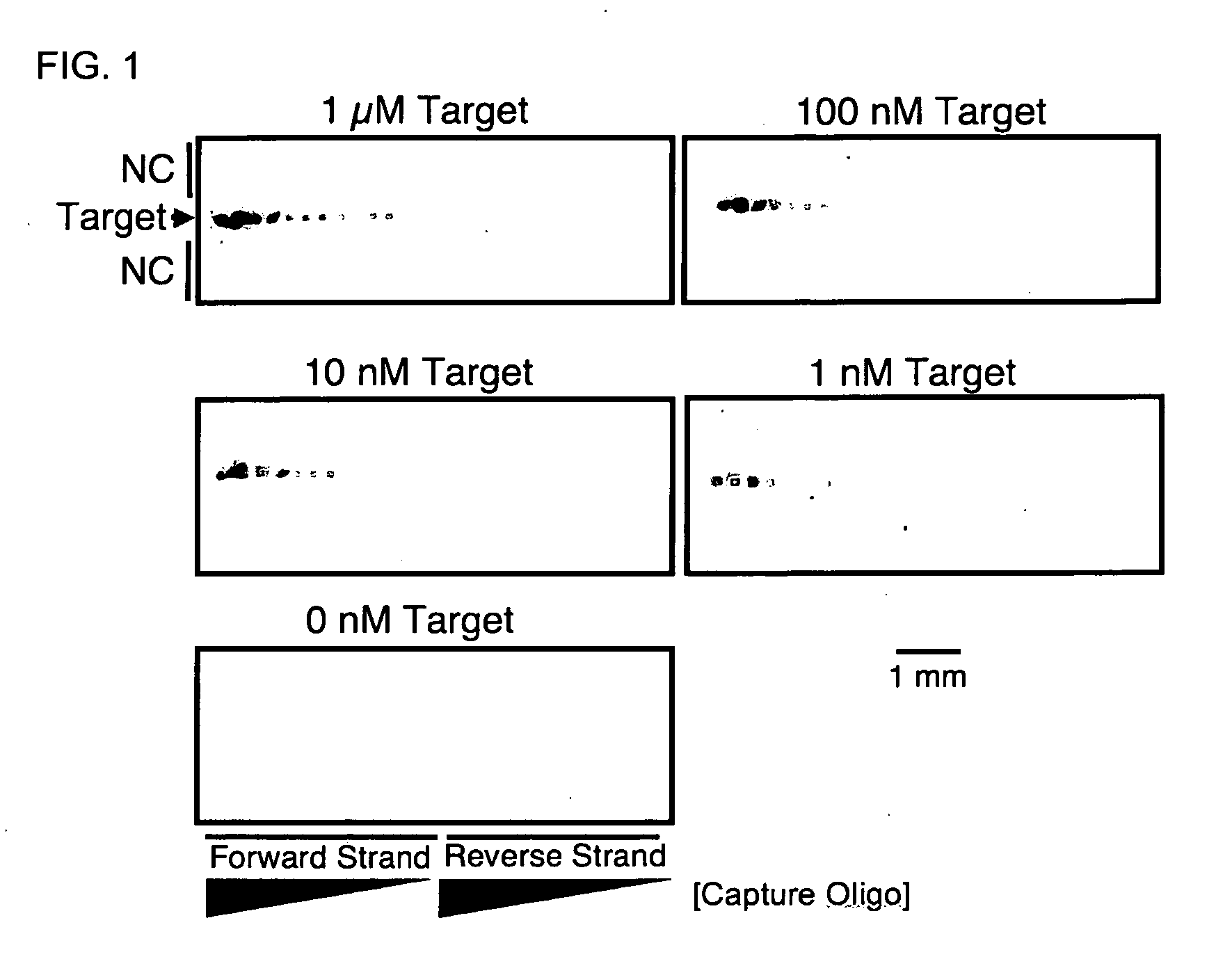
[0097]DNA dipstick microarrays were fabricated at a density of 36 features per mm2 using varying concentrations of capture oligonucleotide, as indicated In FIG. 1. Printing solutions of capture oligonucleotide at 200 μM, 100 μM, 50 μM, 25 μM, 12.5 μM, 6.25 μM, and 3.125 μM were prepared and patterned on to lateral flow membranes. The resulting DNA dipstick microarrays were introduced to 100 μl of synthetic target DNA at the indicated concentration of 1 μM, 100 nM, 10 nM, 1 nM, and 0 nM corresponding to 100 μmol, 10 μmol, 1 μmol, 100 fmol, and 0 fmol target molecules respectively. Capture of 100 fmol target molecule was apparent at capture oligonucleotide printing concentrations as low as 12.5 μM (FIG. 1). However, the most sensitive detection was obtained at higher capture oligonucleotide printing concentrations of 200 μM. Subsequent DNA dipsticks and DNA dipstick micr...
example 2
Detection of Single and Multiple SS-DNA Species
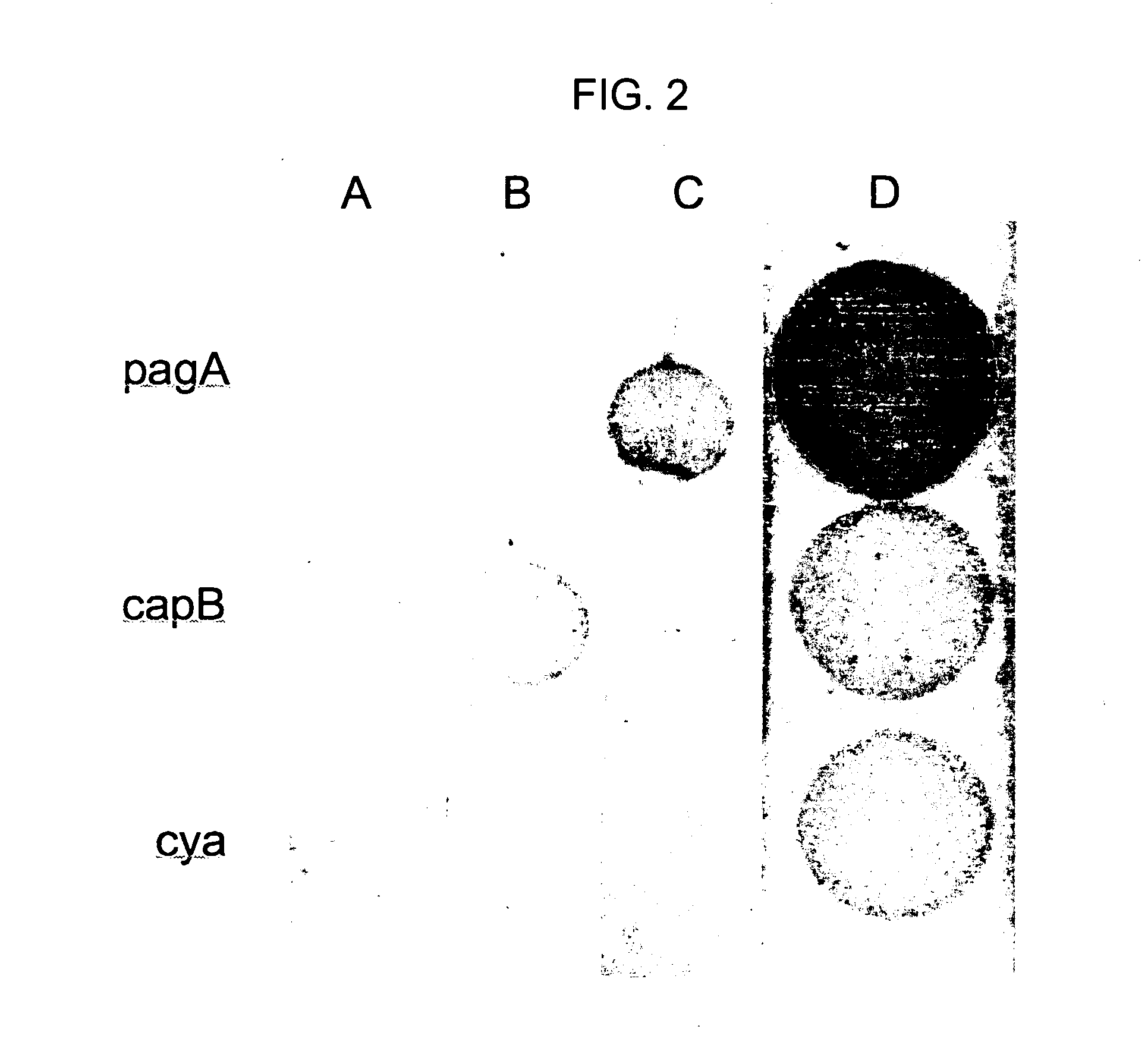
[0098]The following example demonstrates sensitive detection of single-stranded DNAs using hybridization-based capture and dyed-microsphere calorimetric detection.
[0099]Sequences derived from the B. anthracis pagA, capB and cya genes were used to demonstrate multiplexed detection. DNA Dipsticks were patterned with capture sequences for the detection of fragments of three key virulence factors of B. anthracis.
[0100]The capture sequences used were:
pagD:[SEQ ID NO: 7]5′- GCAGGATTTAGTAATTCGAATTTTTTTTTTTTTTT-3′;cyaD908:[SEQ ID NO: 8];5′ TGGTACTAAACCTGAAGCTTTTTTTTTTTTTTTTT 3′;andcapD:[SEQ ID NO: 9]5′-TACATGGTCTTCCCAGATAATTTTTTTTTTTTTTT-3′.
0.35 μm dyed COOH microspheres (Spherotech, Inc.) were coupled using EDAC and standard protocols to:
[SEQ ID NO: 10]5′ amine-C12-TTTTTTTTTTTTTTTTTTCAGAAGAATTCTTACGAAAATTTGAT 3′ for capB detection,[SEQ ID NO: 11]5′ amine-C12-TTTTTTTTTTTTTTTTTCTTTGATATTGGTGGGAGTGTATC for pagA detection;and[SEQ ID NO: 12]5′ ami...
example 3
Sensitivity and Detection Time for DNA Dipstick and DNA Dipstick Microarray
[0102]The sensitivity and time required to detect nucleic acids on DNA dipsticks and DNA dipstick microarrays were evaluated using synthetic target molecules for which exact concentrations could be determined.
[0103]Dipsticks were fabricated by manual deposition of capture oligonucleotides onto membrane strips of approximately 160 to 275 mm2 surface area, features sizes were ˜2-3 mm in diameter. Dipstick microarrays were printed using microarray fabrication robotics to pattern membrane strips of approximately 60 mm2 surface area such that feature sizes were 300-600 μm in diameter. DNA dipsticks were challenged with 400 μl of synthetic target molecule in the presence of appropriate detection microspheres. Typical lateral flow time for these strips was approximately 45 minutes from sample introduction to complete transport of the sample through the dipstick matrix. Dilution series experiments revealed the sensit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
| Pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com