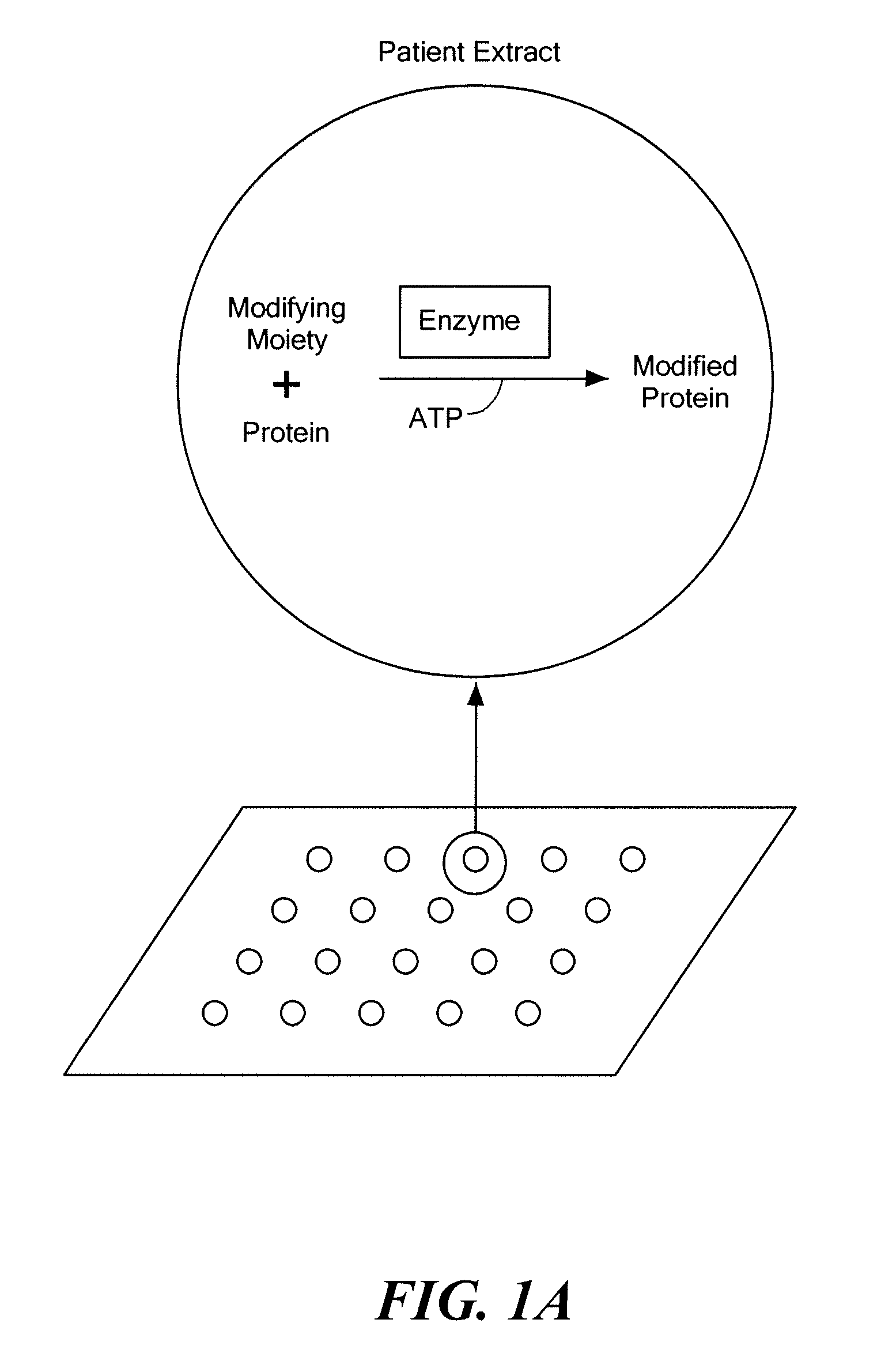
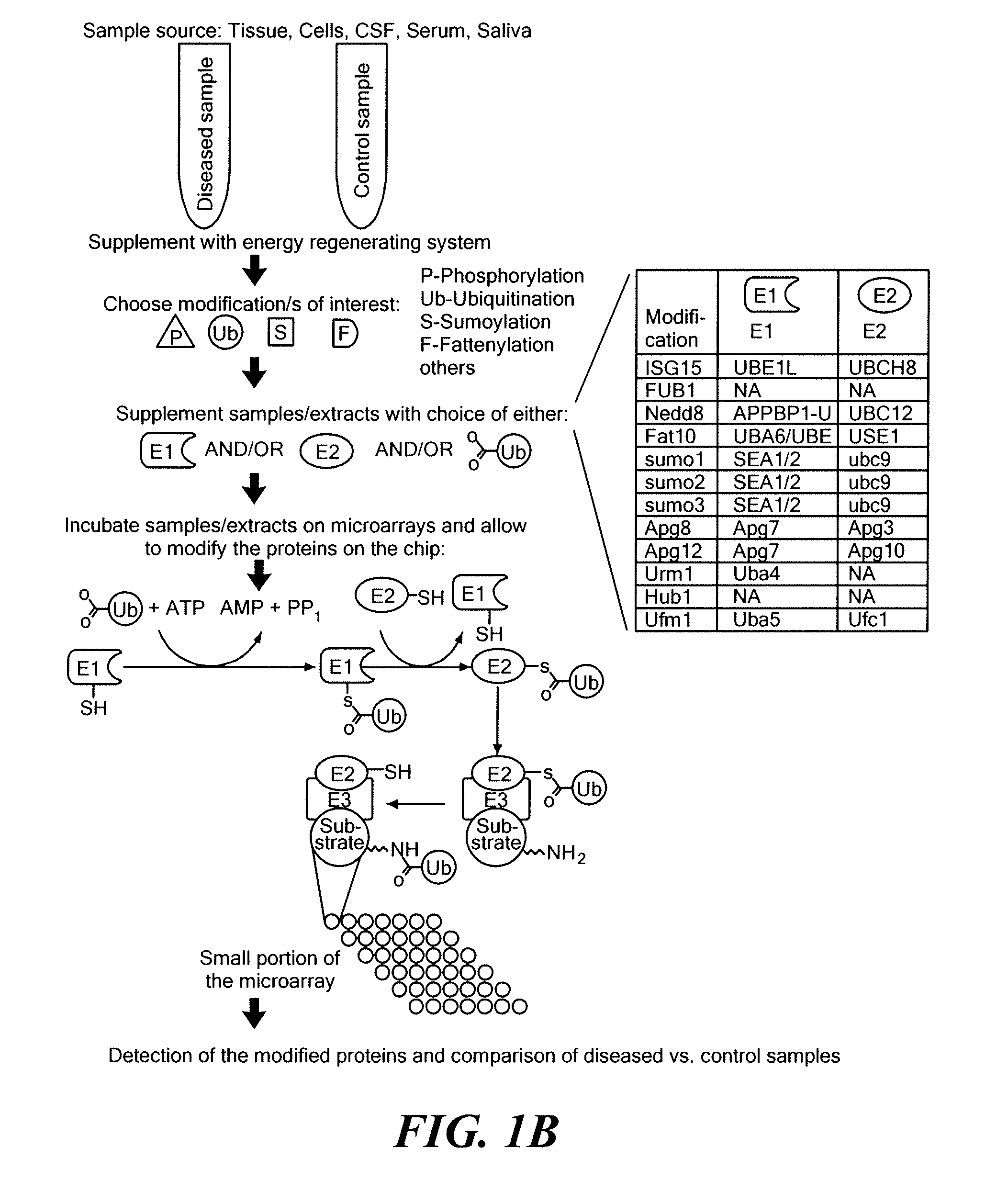
Diagnostic method based on large scale identification of post-translational modification of proteins
a protein and large-scale technology, applied in the field of large-scale protein identification and diagnostic methods, can solve the problems of blot failure to work well, protein identification difficult or impossible, and theoretically and empirically problematic analysis of protein ptm in cell extracts as well as extracellular fluids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
Protein Ubiquitination Patterns Upon Escape from the Spindle Assembly Checkpoint in Mammalian Cells
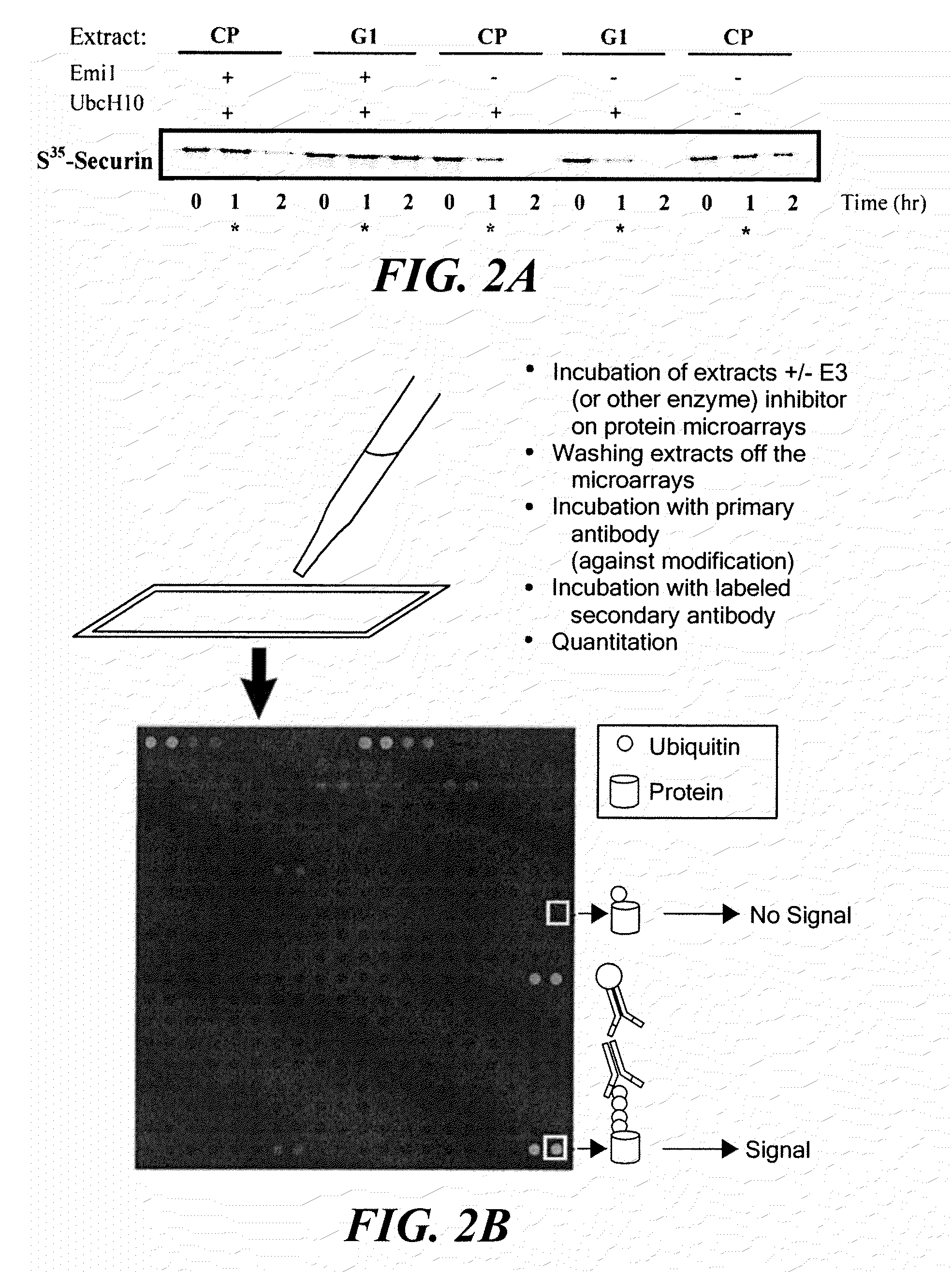
[0150]Protein microarrays were used to identify the polyubiquitination state of proteins under specific cellular conditions. Highly concentrated cellular extracts that have demonstrable function specific for a particular phase of the cell cycle were used to modify the polyubiquitination state of human proteins on a microarray.
[0151]Specifically, the degradation of proteins involved in mitosis was examined by determining the polyubiquitination state of certain proteins at specific stages of the cell cycle. During mitosis, rapid degradation of the mitotic cyclins (11, 12) causes abrupt shut-down of mitotic kinase activity, allowing the cell to enter anaphase. The Anaphase Promoting Complex (APC), a multi-subunit E3 ligase, targets cyclins and other mitotic substrates for proteasomal degradation (13, 14) which in turn leads to the metaphase to anaphase transition. Thus, cell division is h...
example ii
Ubiquitination of Human Brain Proteins in Alzheimer's Disease
[0165]Human brain specimens are collected from deceased human subjects at autopsy after obtaining informed consent from the next of kin under protocols approved by the Partners Human Research Committee at Brigham and Women's Hospital. Weighed frozen human temporal or frontal cortices containing white and gray matter are added to freshly prepared, ice-cold TBS (20 mM Tris-HCl, 150 mM NaCl, pH 7.4) at a ratio of 4:1 (TBS volume / brain wet weight) and homogenized with 25 strokes at a setting of 10 on a mechanical Dounce homogenizer. The homogenate is centrifuged at 175,000×g in a TLA100.2 rotor on a Beckman TL 100 centrifuge, and then the supernatant is aliquoted and stored at −80° C.
[0166]For analysis of ubiquitination, samples are thawed on ice, supplemented with 5 μM ubiquitin, 2 mM ATP, and 150 mM creatine phosphate, and then incubated on a microarray to carry out the ubiquitination reaction. Optionally, E1 and / or E2 enzym...
example iii
Protein Ubiquitination in Cerebrospinal Fluid (CSF) from a Patient with Brain Tumor
[0167]Undiluted CSF from a patient with brain tumor was analyzed for enzyme activity responsible for PTM (ubiquitination) of human proteins. Conditions were similar to conditions used for cellular extracts. An ATP regenerating system and ubiquitin were added to the CSF sample, and the mixture was reacted with a protein microarray containing 8000 human proteins. A control reaction contained the same CSF sample but was not supplemented with ubiquitin or the energy mix.
[0168]A specific subset of proteins that are disproportionately expressed in brain (compared to a background of all the proteins that were on the chip) were found to be ubiquitinated (i.e., showed at least 2.5-fold higher signal than in the control), as shown in FIG. 11. The proteins that underwent CSF-mediated ubiquitination were distinct from background modification seen under control conditions. The functional annotation categories (gen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com