Discovery and validation of cancer biomarkers using a protein analysis methodology to analyze specimens
a protein analysis and specimen technology, applied in the field of gene and proteomics, can solve the problems of cell death, inability to detect cancer biomarkers, and inability to detect cancer biomarkers, and achieve the effect of optimizing the selection of therapeutic agents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0075]In order to accurately measure oncoprotein expression and activation in limited clinical specimens, we have developed a nano-fluidic proteomic immunoassay detection method (NIA) that combines isoelectric protein focusing and antibody detection. Here we demonstrate that we can use this new technique to quantitate oncoprotein expression and phosphorylation in clinical specimens to precisely measure specific changes in phospho-isomers of oncoproteins in vitro and in vivo.
Results
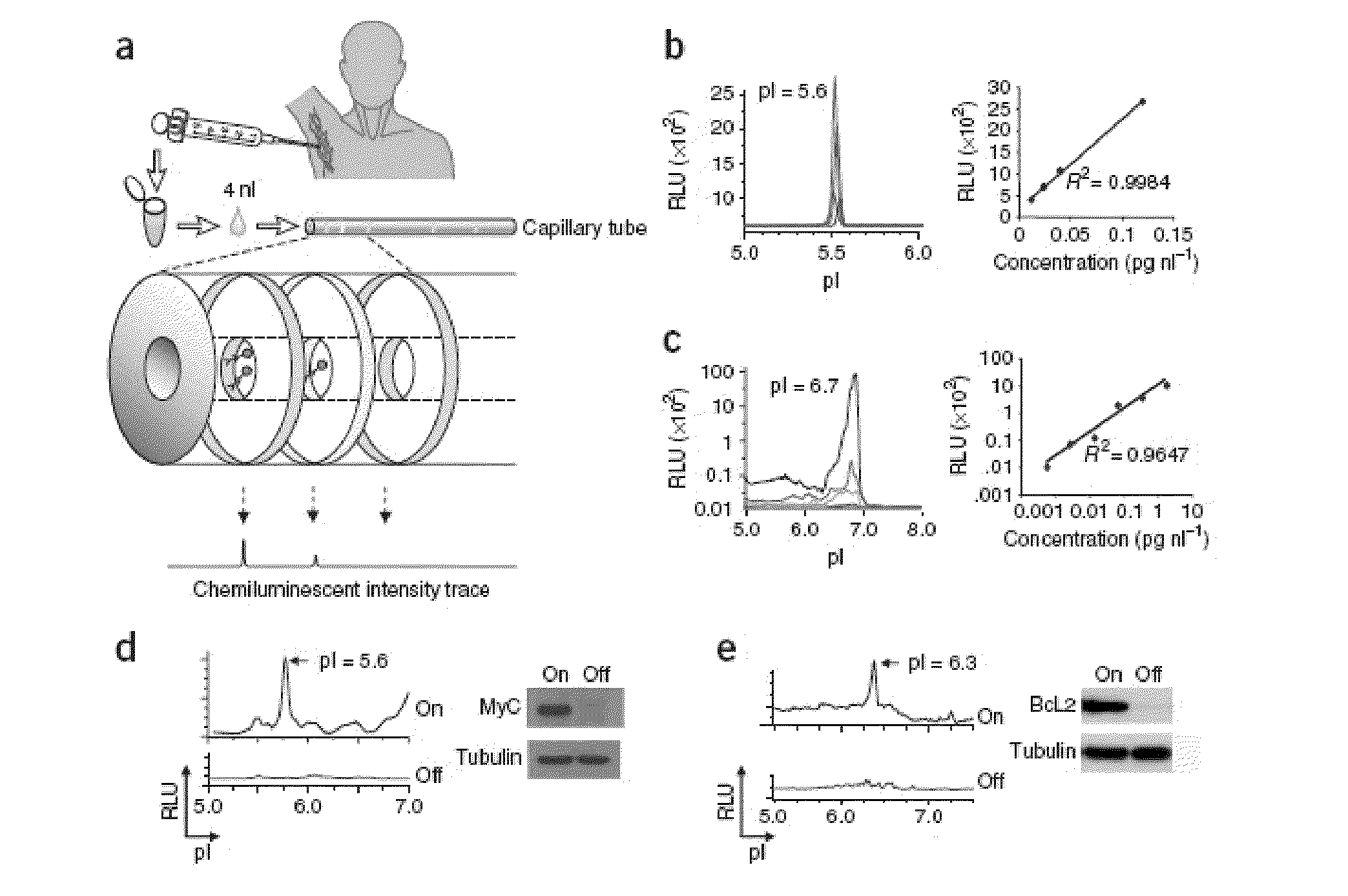
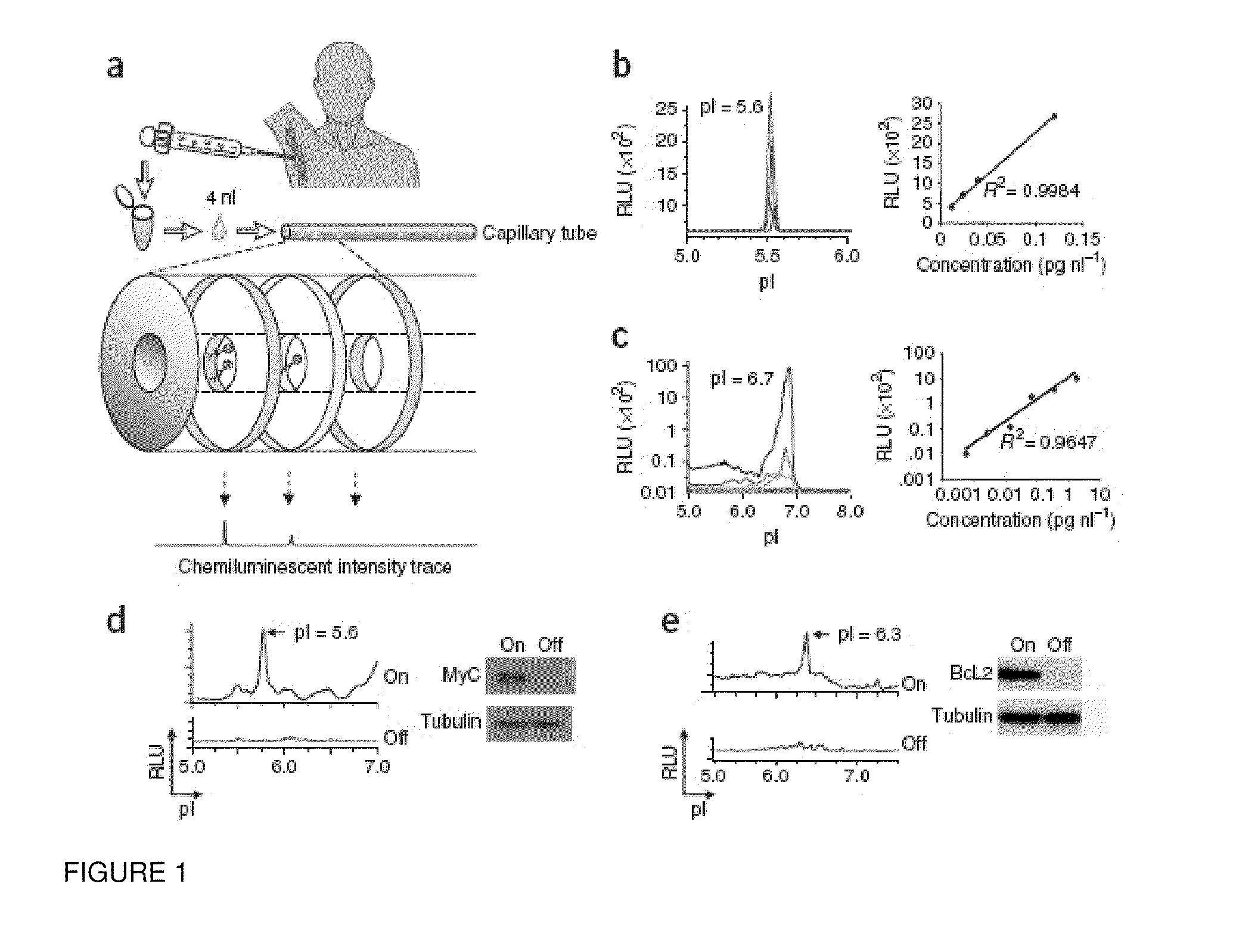
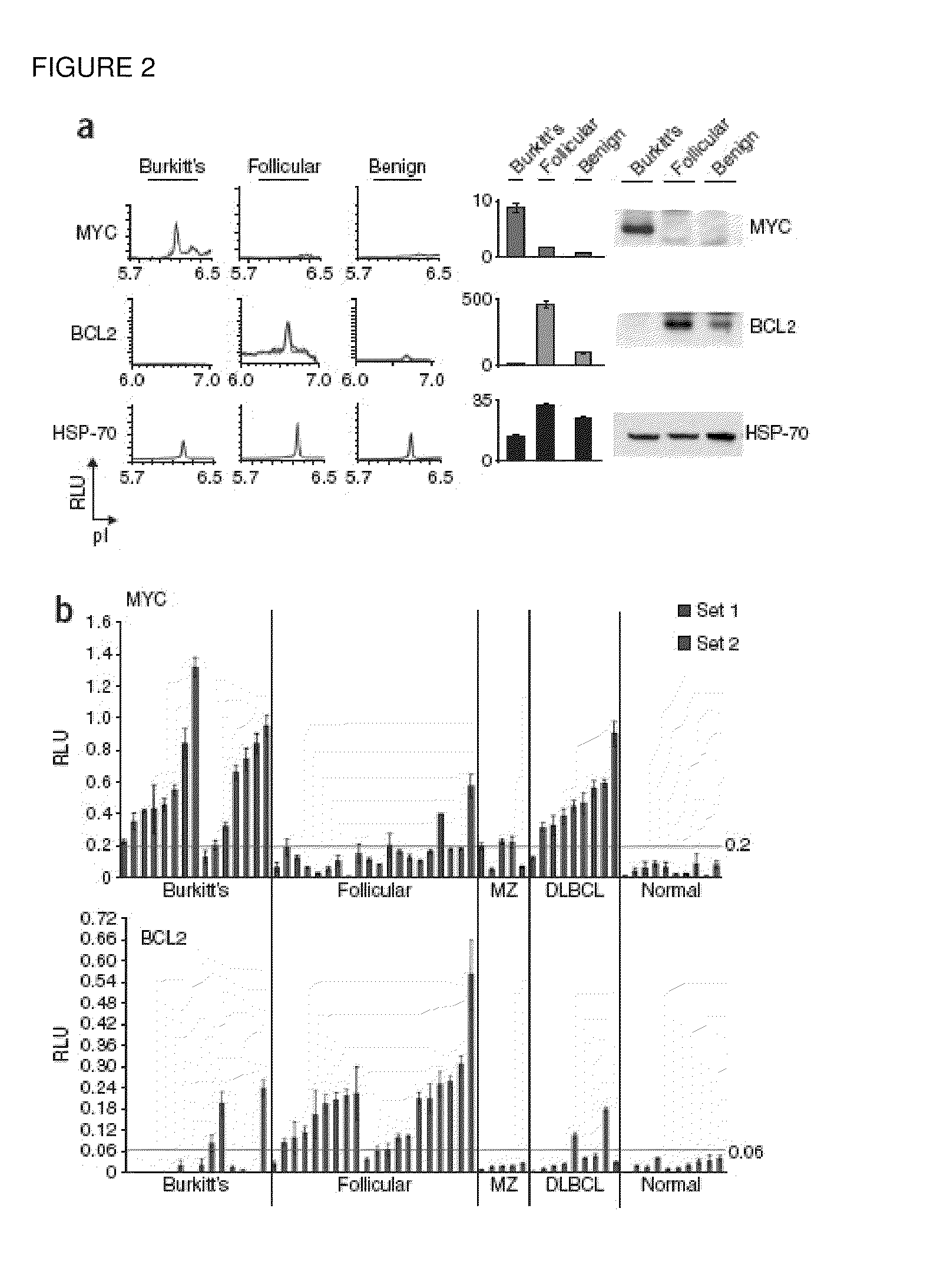
[0076]NIA Detection of Oncoprotein Expression in Clinical Specimens. NIA incorporates isoelectric focusing of proteins, followed by antibody detection of specific epitopes with chemiluminescence. The chemiluminescent signal is rendered as a chemiluminescence isoelectropherogram (trace) of “relative luminescence units (RLU)” on the y-axis vs. isoelectric point (pI) on the x-axis (FIG. 1a). As little as 2 picograms of MYC could be detected using 4 nanoliters of recombinant protein per capillary (final capill...
example 2
Nanoscale Quantification of Phosphorylated and Unphosphorylated ERK and MEK Isoforms Differentiates Tumor and Non-Tumor Clinical Specimens
[0108]We have developed the use of a highly sensitive microfluidic nano-immunoassay system (NIA) to perform detailed analysis of ERK and MEK activation in hematopoietic and solid tumors. We described above the use of NIA for measurement of proteins in as little as 4 nL of lysate from lymphoma and leukemia specimens. Now, we present results measuring specific isoforms of MAPK proteins in fine needle aspirates (FNAs) from patients with solid tumors and in blood buffy coats from patients with Myelo-Dysplastic Syndrome (MDS).
[0109]Using a single antibody that recognizes both the phosphorylated and unphosphorylated isoforms of ERK, we can determine levels of each ERK isoform, and also percent phosphorylation of ERK. NIA revealed that different tumor types could be distinguished based upon differing patterns of ERK isoforms. To determine if NIA can meas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com