Method of Preparing Piceatannol Using Bacterial Cytochrome P450 and Composition Therefor
a technology of cytochrome p450 and piceatannol, which is applied in the field of preparation of piceatannol using bacterial cytochrome p450 and composition therefor, can solve the problems of large quantities of metabolites to be produced, inability to meet the requirements of biocatalysis, and difficulty in synthesis of pure metabolites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of P450 BM3 Mutants By Site-Directed Mutagenesis
[0051]Site-directed mutants of CYP102A1 were prepared as disclosed by Kim et al., Drug Metab Dispos, volume 35, p. 2166-2170, 2008. PCR primers used to introduce BamHI / SacI restriction sites and to induce mutation are listed in Table 1. Codons for amino acid substitution are in italics and underlined. The PCR primers were obtained from Genotech (Daejeon, Korea). Genes that encode CYP102A1 mutants were amplified from pCWBM3 by PCR using primers designed to facilitate cloning into an expression vector pCWori (Dr. F. W. Dahlquist, University of California, Santa Barbara, Calif.) or pSE420 (Invitrogen) (Farinas et al., 2001). Oligonucleotide assembly was performed by PCR using the 14 sets of designed primers listed in Table 1. The amplified genes were subsequently cloned into the PCWBM3 BamHI / SacI vector at the BamHI / SacI restriction sites. These plasmids were transformed into Escherichia coli DH5a F′IQ (Invitrogen), and this ...
example 2
Expression And Purification of Wild-Type CYP102A1 And Mutants of CYP102A1
[0052]Plasmids comprising a gene of wild-type CYP102A1 and mutants of CYP102A1 (pCWBM3) were transformed into Escherichia coli DH5α F′-IQ. A single colony was inoculated into 5 ml of a Luria-Bertani medium supplemented with ampicillin (100 g / ml) and cultured at 37° C. This culture was inoculated into 250 ml of a Terrific Broth medium supplemented with ampicillin (100 g / ml). The cells were grown at 37° C. while shaking at 250 rpm to an 0D600 of up to 0.8, at which gene expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 0.5 mM. δ-Aminolevulinic acid (1.0 mM) was also added thereto. Following induction, the cultures were allowed to grow another 36 hours at 30° C. Cells were harvested by centrifugation (15 min, 5000 g, 4° C.). The cell pellet was resuspended in a TES buffer (100 mM Tris-HCl, pH 7.6, 500 mM sucrose, 0.5 mM EDTA) and lysed by sonication (Sonicato...
example 3
Hydroxylation of Trans-Resveratrol By Wild-Type P450 BM3 And Mutants of P450 BM3
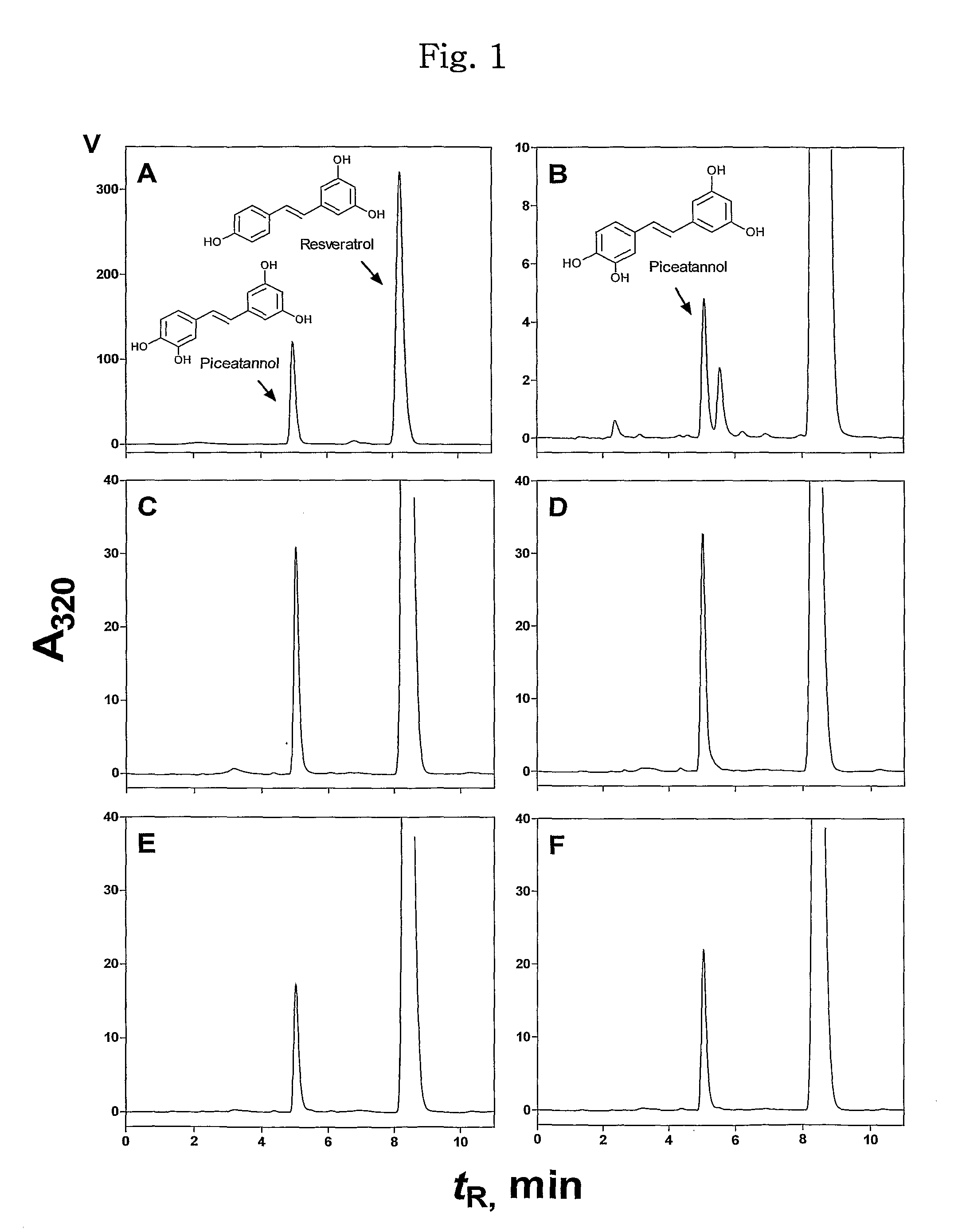
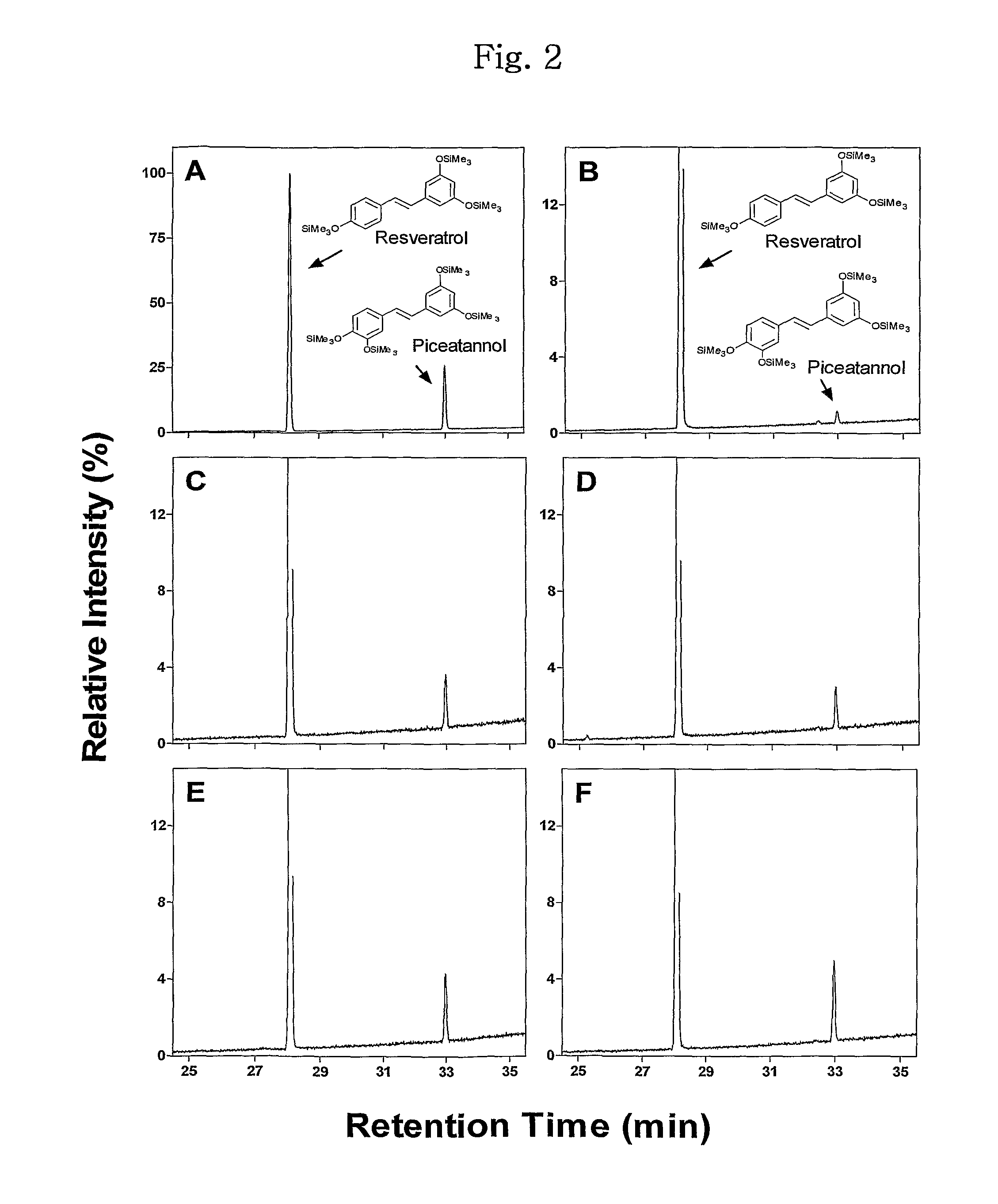
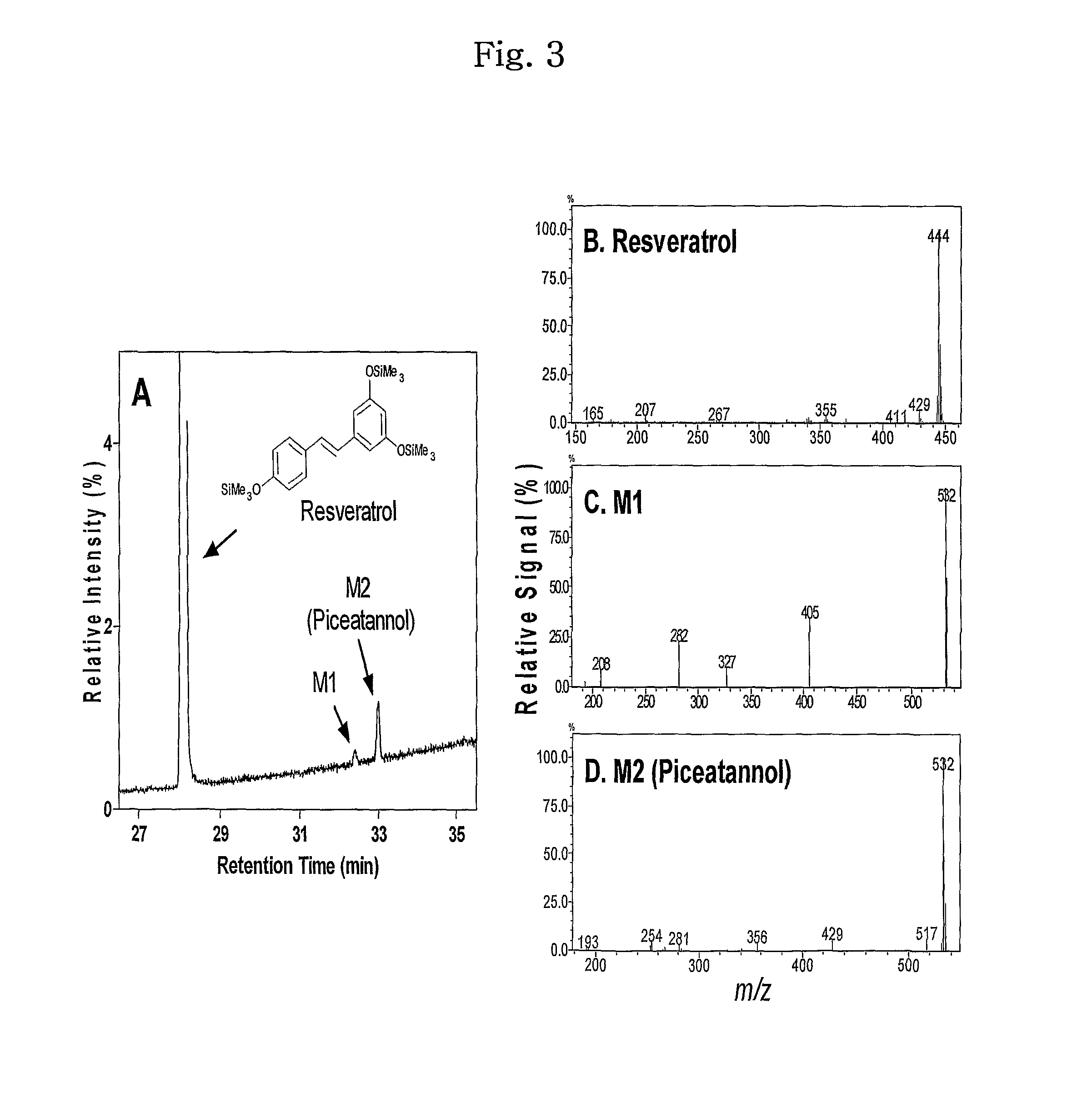
[0055]Oxidation of trans-resveratrol, a substrate of human CYP1A2, by CYP102A1 was identified. Typical steady-state reactions for trans-resveratrol hydroxylation included 50 pmol P450 BM3 in 0.25 ml of a 100 mM potassium phosphate buffer (pH 7.4) were performed along with a specified amount of a substrate. To determine the kinetic parameter of several CYP102A1 mutants, 2 to 100 μM of trans-resveratrol was used. An NADPH-generating system was used to initiate reaction solutions (final concentrations: 10 mM glucose 6-phosphate, 0.5 mM NADP+, and 1 IU yeast glucose 6-phosphate per ml). Trans-resveratrol stocks (20 mM) were prepared in DMSO and diluted into the enzyme reactions with the final organic solvent concentration <1% (v / v). Reactions were generally incubated for 10 min at 37° C., and terminated with 105 μl of ice-cold acetic acid / methanol (95 / 5, v / v).
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com