Nanoscale imaging of molecular positions and anisotropies
a technology of anisotropies and molecular positions, applied in the field of system and method of imaging at a nanoscale level, can solve the problems of limiting the resolution to 150-250 nm, not providing information about anisotropy and rotational freedom of individual molecules, and achieves improved spatial resolution, improved p-fpalm resolution, and preferential orientation of molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
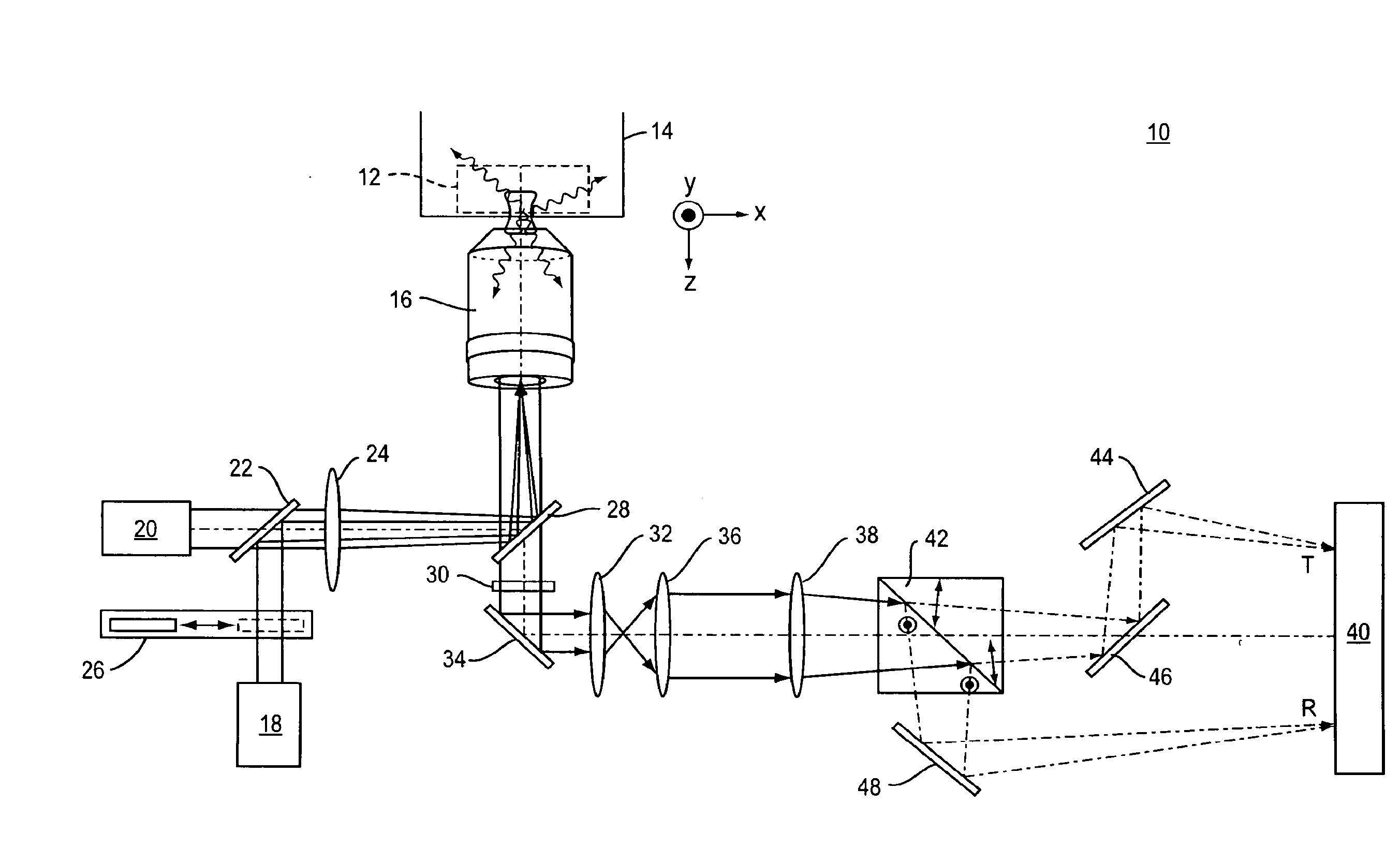
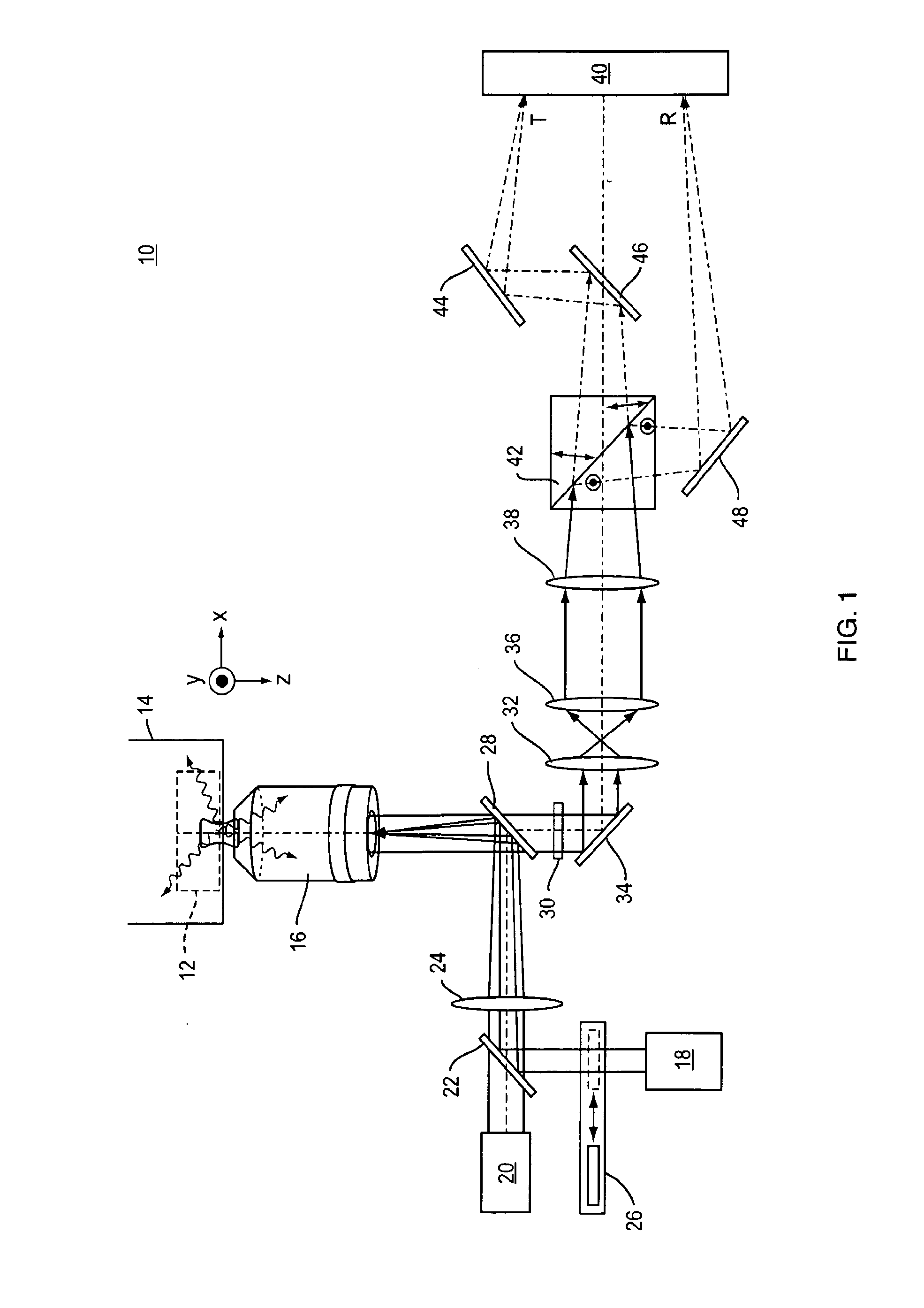
[0038]Referring to FIG. 1, a representative example of an imaging system 10 of the present invention includes a standard two-dimensional Fluorescence Photoactivation Localization Microscopy (FPALM) system with a modified detection path. A sample 12 labeled with a suitable fluorophore is placed on the stage 14 of any suitable microscope together with a suitable imaging lens 16. One representative but non-limiting example of a suitable microscope is the Olympus IX-71 inverted microscope (Olympus America, Melville, N.Y.) with a 60×1.2 NA water-immersion objective (UPLAPO60XW, Olympus) as the imaging lens. The use of a water-immersion lens is beneficial because it minimizes aberrations when imaging a sample that is also in water, as is the case with many biological samples. High numerical aperture lenses and other types of immersion lenses such as oil-immersion, glycerol-immersion, and air immersion lenses are also well-suited for use in the present application.
[0039]The sample 12 is il...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com