Engineering the pathway for succinate production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of KJ073 Strain for Succinate Production
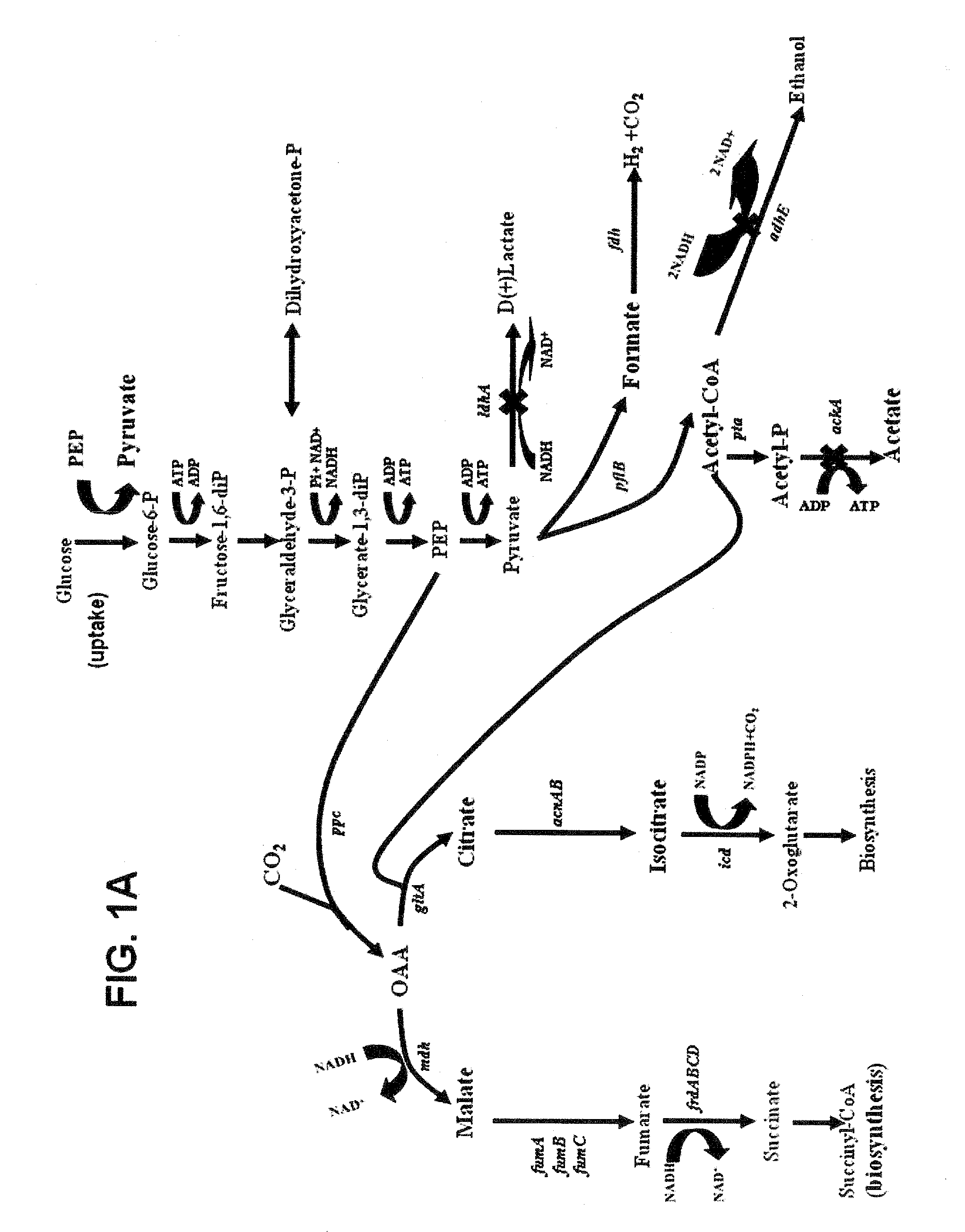
[0146]In the construction of a strain suitable for succinic acid production in minimal medium containing 5% glucose, both a rational design approach and metabolic evolution were followed. By inspection of FIG. 1 illustrating the generally accepted standard fermentation pathways for E. coli, a rational design for the metabolic engineering of strains producing succinate was devised in which insertional inactivations were made in genes encoding the terminal steps for all alternative products: lactate (ldhA), ethanol (adhE) and acetate (ackA). Results from this metabolic engineering by rational design were completely unexpected. The KJ012 (ΔldhA::FRT ΔadhE::FRT ΔackA::FRT) strain resulting from the insertional inactivation of ldhA, adhE and ack genes grew very poorly under anaerobic conditions in mineral salts medium containing 5% glucose (278 mM) and produced acetate instead of succinate as the primary fermentation product. Counter t...
example 2
Construction of KJ134 Strain for Succinate Production
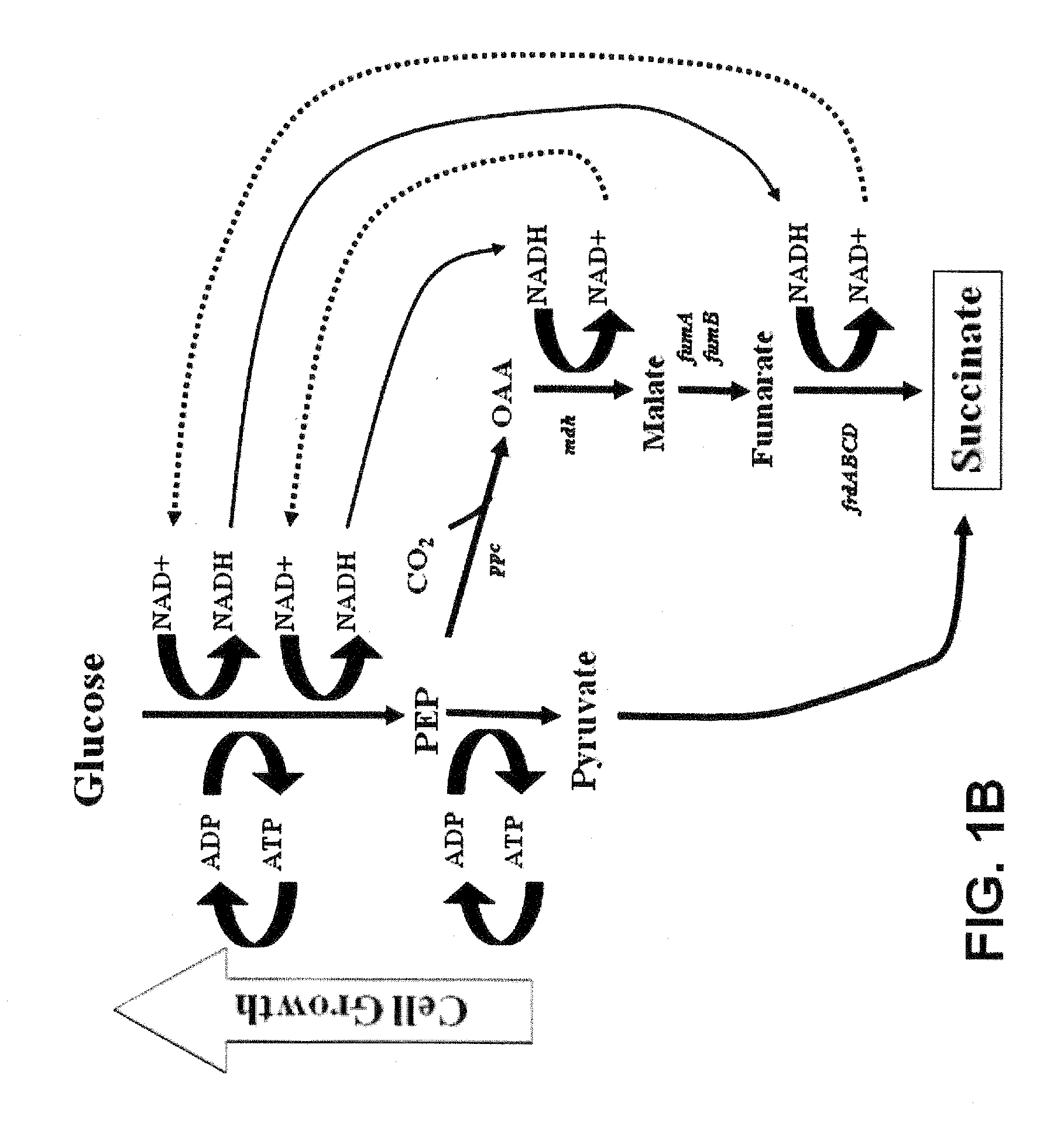
[0173]As described in the Example 1 above, the central anaerobic fermentation genes in E. coli C wild type were sequentially deleted by the strategy of Datsenko & Wanner (2000) with PCR products and removable antibiotic markers (by using FRT recognition sites and FLP recombinase). These constructions in combination with metabolic evolution (growth-based selection for increased efficiency of ATP production) were used to select for a mutant strain that recruited the energy-conserving, phosphoenolpyruvate carboxykinase (pck) to increase growth and succinate production (FIG. 9). The resulting strain, KJ073, produced 1.2 moles of succinate per mole of metabolized glucose (Jantama et al., 2008a) and uses a succinate pathway quite analogous to the rumen bacteria, Actinobacillus succinogenes (van der Werf et al., 1997) and Mannheimia succiniciproducens (Song et al., 2007). However, methods used to construct these gene deletions left a sin...
example 3
Recruitment of Gluconeogenic PEP Carboxykinase (pck) for Succinate Production
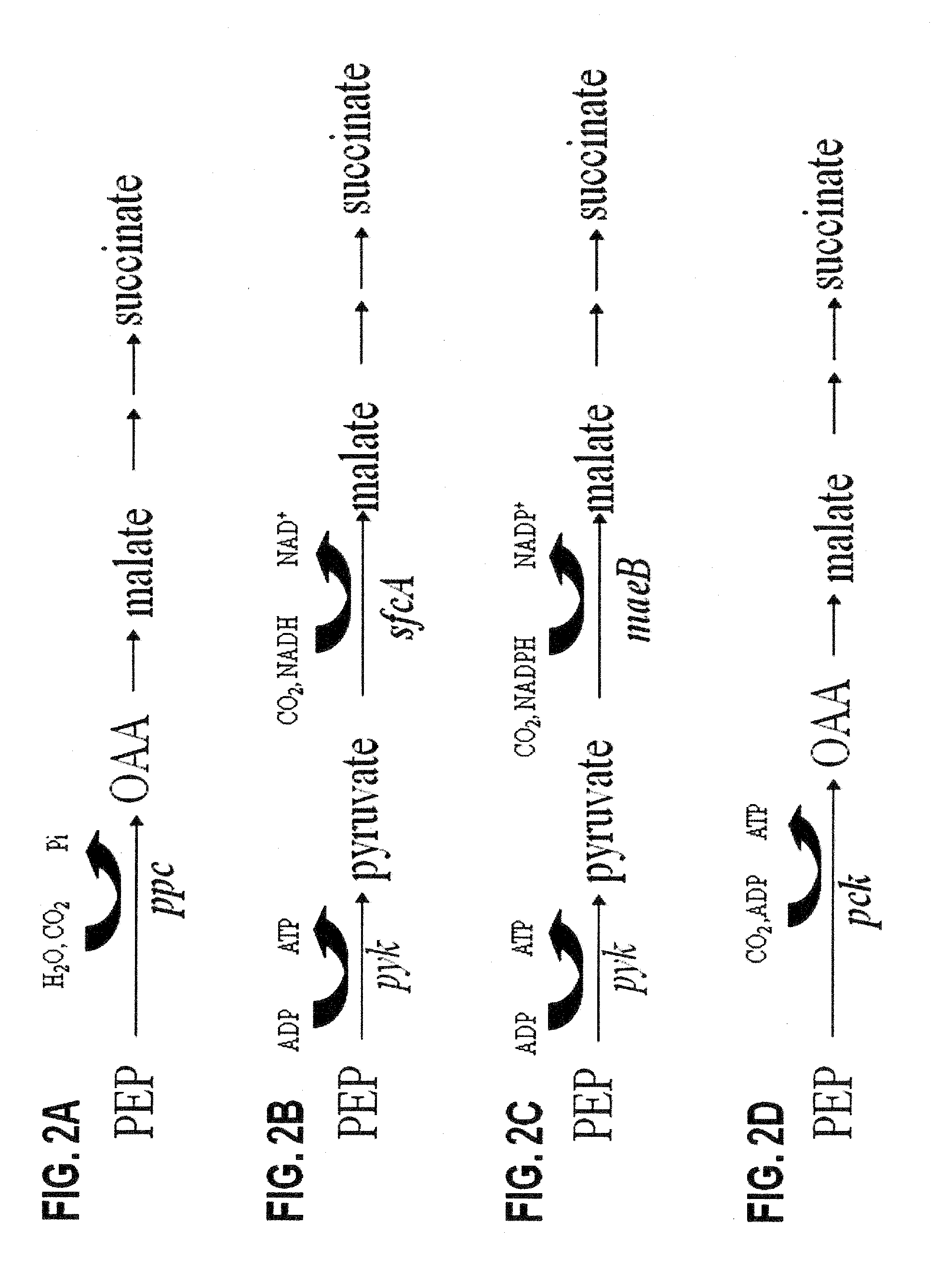
[0184]E. coli has the metabolic potential for four native carboxylation pathways that could be used to produce succinate (FIGS. 2A-2D). The carboxylation of phosphoenolpyruvate (PEP) to oxaloacetate (OAA) by phosphoenolpyruvate carboxylase (ppc) is recognized as the primary pathway for the fermentative production of succinate in E. coli (FIG. 2A). (Fraenkel DG, 1996; Unden & Kleefeld, 2004; Karp et al., 2007). This reaction is essentially irreversible due to the energy loss associated with the release of inorganic phosphate. The three other carboxylation reactions are normally repressed by high concentration of glucose in the medium and also reported to function in the reverse direction for gluconeogenesis. (Samuelov et al., 1991; Oh et al., 2002; Stols & Donnelly, 1997). The second and third carboxylation pathways (FIGS. 2B and 2C) use NADH-linked and NAPDH-linked malic enzymes (sfcA and maeB), respectivel...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com