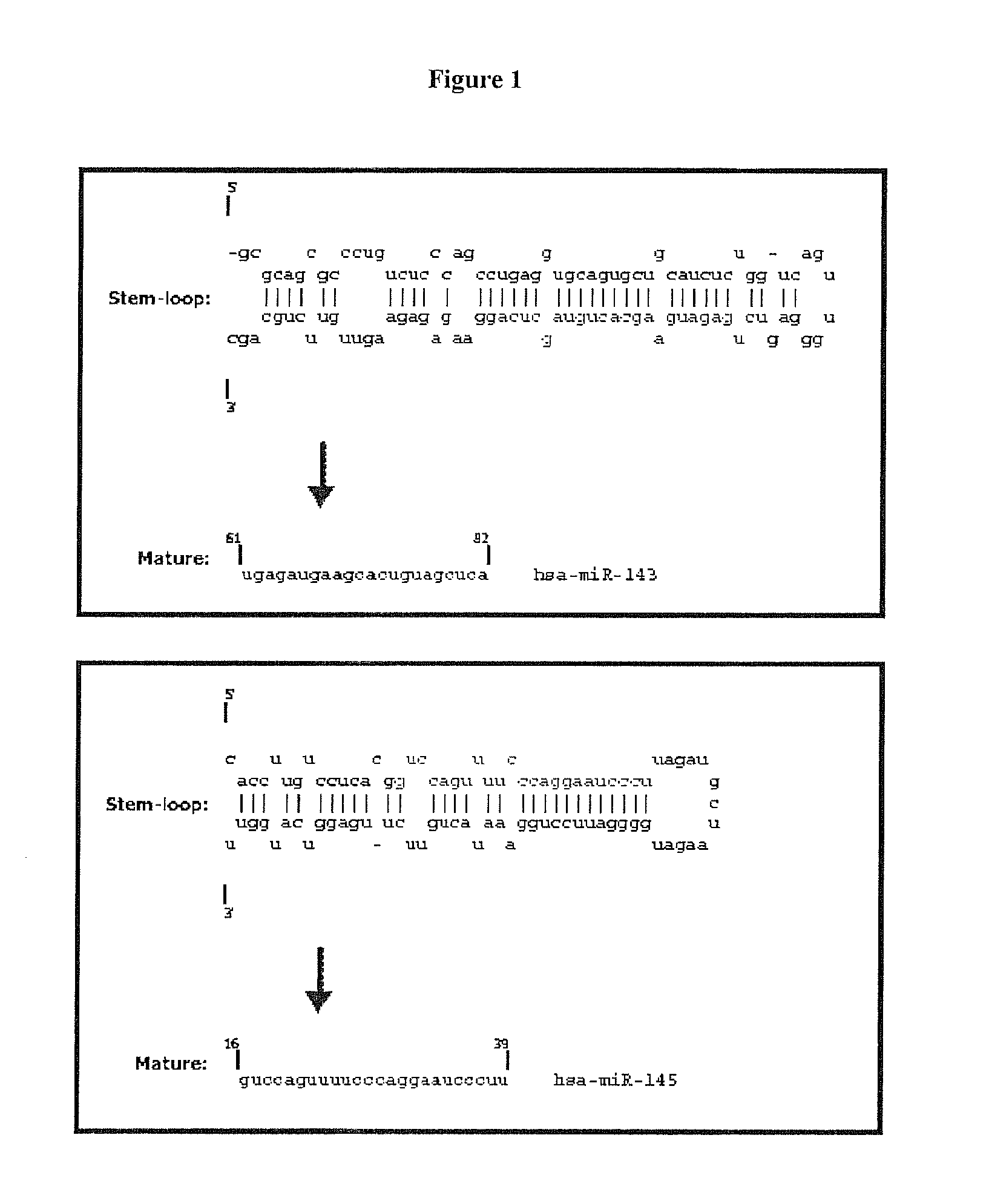
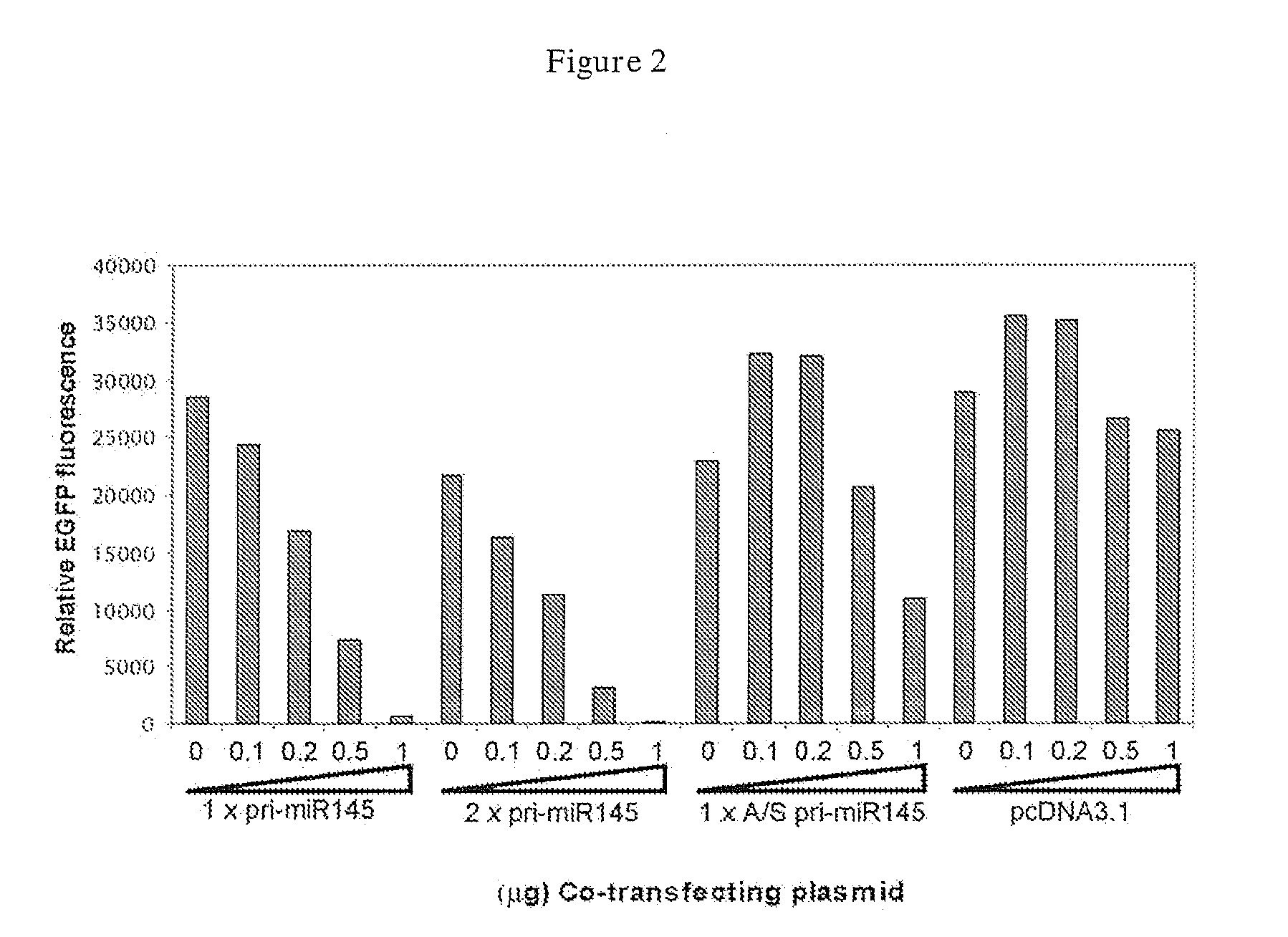
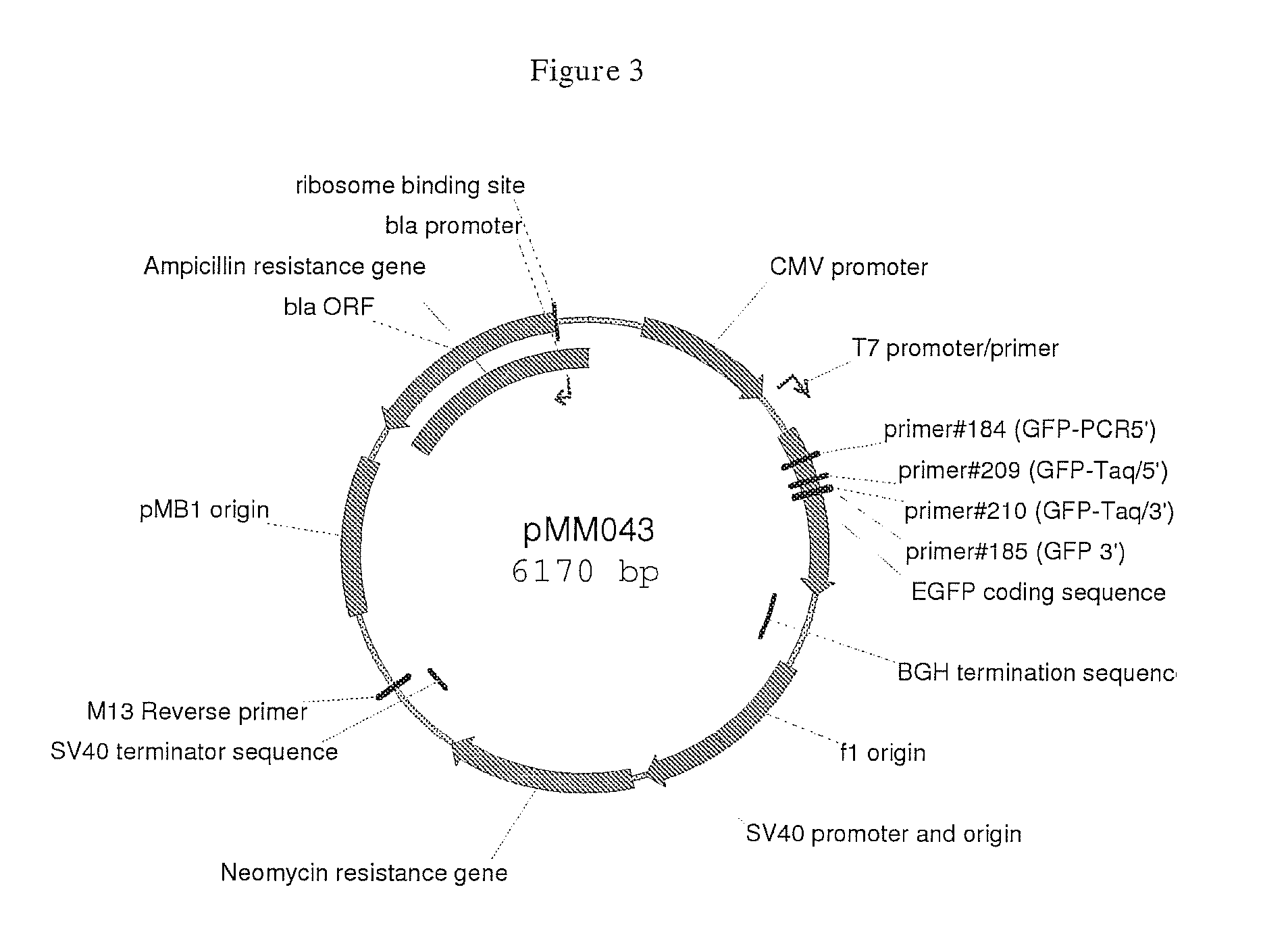
Targeting cells with altered microrna expression
a technology of microrna and target cells, which is applied in the direction of nerve system cells, drug compositions, line-transmission, etc., can solve the problems of inability to restrict the expression of the delivered gene to the desired tissue or type of cell, and inability to control the expression of the therapeutic gene, so as to improve cell survival or rescue, improve cell proliferation, and modulate the development of cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNA Isolation From Tissue Samples and Cell Lines
[0234]Cell lines may be maintained in an appropriate medium. Colorectal tumors and the corresponding normal mucosae may be obtained from fresh surgical resections, following informed consent from patients, and then classified according to standard histopathological classification methods.
[0235]RNA may isolated from cell lines, using Trizol reagent (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. RNA may be purified from colorectal tissues using the procedure of Chomczynski, P. and Sacchi, N. (1987). Anal. Biochem. 162: 156-159.
example 2
Cloning of MicroRNAs miRNAs may be cloned essentially as described by Elbashir et al. (2001) Genes Dev. 15: 188-200, except that nucleic acids may be electroeluted from acrylamide gel slices using the Biotrap system (Schleicher and Schuell GmbH, Dassel, Germany). Briefly, small RNA fractions of between 18 and 26 bases may be size selected on a denaturing polyacrylamide gel. Adapter oligonucleotides, containing EcoRI restriction sites, may then be directionally ligated to the RNA molecules. The adapter-ligated RNA may then be amplified by RT-PCR. Concatamerized fragments, containing multimers of religated, EcoRI-digested PCR products, between 200 and 650 bp, are size selected on an agarose gel and recovered by electroelution. The concatamers may then be end-repaired and dA-tailed with Taq DNA polymerase, then cloned into pGEM T-easy (Promega, Madison, Wis.) or pTOPO (Invitrogen) according to the manufacturers' instructions. Plasmid inserts from the resultant colonies may be analyzed ...
example 3
Northern Analysis
[0236]Total RNA (20 μg) may be separated on a 15% denaturing polyacrylamide gel. Loadings are visualized by ethidium bromide staining. The RNA may then be transferred to Hybond N+ nylon membrane by semi-dry blotting (OWL Separation Systems, Portsmouth, N.H.). Probes may be generated by T4 Polynucleotide Kinase (New England Biolabs, Beverly, Mass.) mediated end-labeling of DNA oligonucleotides with [γ-32P]ATP. To increase the specific activity of the probes, the miRNA sequence may be concatamerized as a trimer of direct repeats, then cloned into pGEM T-easy and the insert amplified using PCR with M13 forward and reverse primers. Antisense probes may then be synthesized using Taq polymerase-generated linear amplification from the Sephadex G-50-purified PCR products to incorporate multiple [γ-32P]dCTP bases.
[0237]Filter hybridization may be performed in QuikHyb Solution (Stratagene, La Jolla, Calif.) containing 106 cpm / ml probe for 1 h, with washes, as per the manufact...
PUM
| Property | Measurement | Unit |
|---|---|---|
| real time RT- | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| chemical properties | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com