Polymerase
a polymerase and polymerase technology, applied in the field of dna polymerases, can solve the problems of limiting the use of unnatural or modified nucleotide bases and the applications they enable, the use of polymerase enzymes for direct incorporation of dye labelled nucleotides, and the inability to complete the substitution of every reactive nucleotide, etc., to achieve the effect of reducing discrimination and high detection agent labeling
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
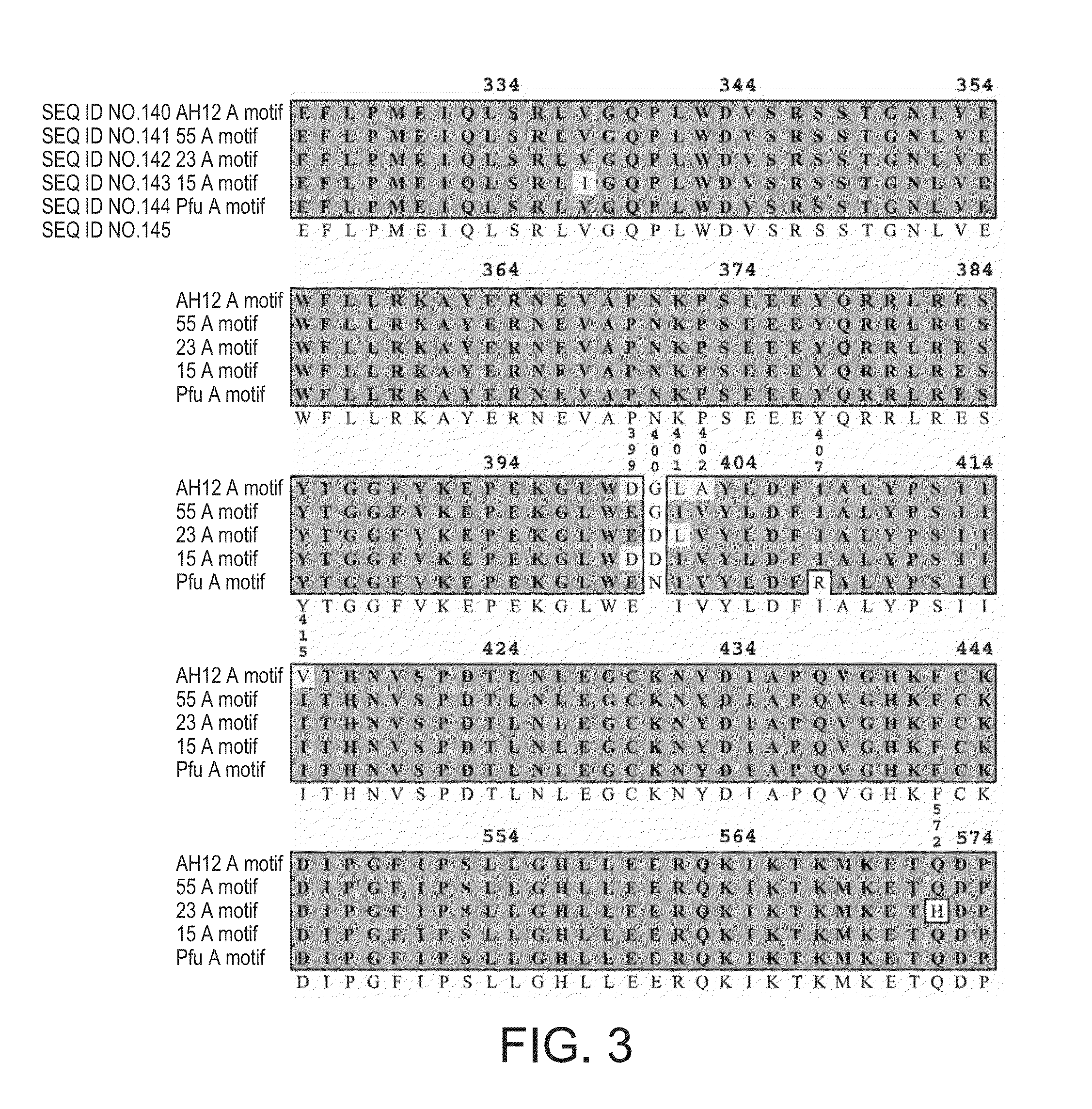
Cloning of Pfu Polymerase Open Reading Frame into pASK
[0204]The Pfu polymerase open reading frame was amplified by PCR from pETpfu (Lu and Erickson, 1997) using primers 1 and 2. The amplified fragment was restriction digested with NdeI and SalI and ligated into pASKDpo4 (Skerra, 1994) from which the NdeI site at 2421 bp had been eliminated from the pASK vector backbone by site-directed-mutagenesis using primers 3 and 4 thereby creating the expression vector pASKpfu.
[0205]The 3″-5″ exonuclease activity of the Pfu enzyme was disabled by site-directed-mutagenesis of exonuclease domain I (Derbyshire et al., 1995) using primers 5 and 6 and pASKpfu as a template thereby generating pASKpfuexo−.
[0206]The XbaI site present in the Pfu sequence at 1683 bp was removed by site-directed-mutagenesis using primers 7 and 8 and pASKpfuexo− as a template thereby generating pASKpfuexo−1.
[0207]To facilitate library construction by iPCR the BsaI site present in the Pfu sequence at 1636 bp was eliminated ...
example 2
Pfu Protein Expression
[0212]Plasmid constructs or libraries were transformed into E. coli Ace6 or TG1TR and expressed as described (Skerra, 1994). Briefly, transformed Ace6 cells are grown overnight at 37° C. in 2×TY, 0.1 mg / mL ampicillin. For expression overnight cultures were diluted 1:50 in 2×TY 0.1 mg / mL ampicillin, grown to an OD595 of 0.6 at 37° C. and induced for protein expression by the addition of anhydrotetracycline. Protein expression was induced for 6 hours at 37° C.
[0213]Cells were harvested by centrifugation, resuspended in 20 ml (per litre of culture) of buffer A (50 mM Tris pH8.0, 1% glucose, 1 mM EDTA), Buffer B (10 mM Tris pH8.0, 50 mM KCl, 1 mM EDTA, 0.5% NP40) was added to a final volume of 50 ml and cells were lysed for 30 min at 75° C. Debris was pelleted by centrifugation and the NaCl was added to the supernatant to 0.25M final concentration. Then neutralized polyethyleneamine (PEI) was added to a final concentration of 0.1% v / v and precipitate pelleted by ce...
example 3
Pfu Requires Modified CSR Conditions
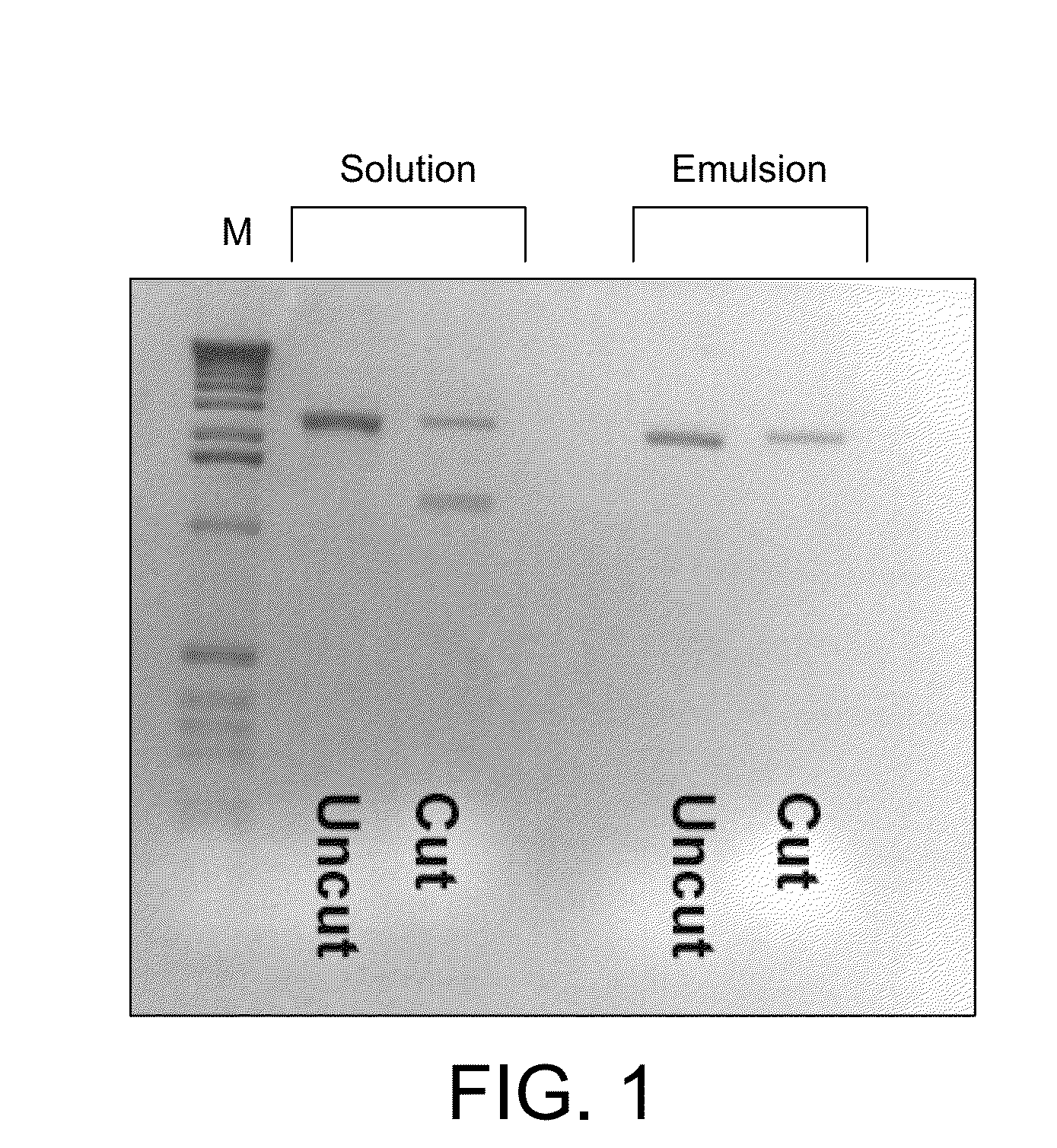
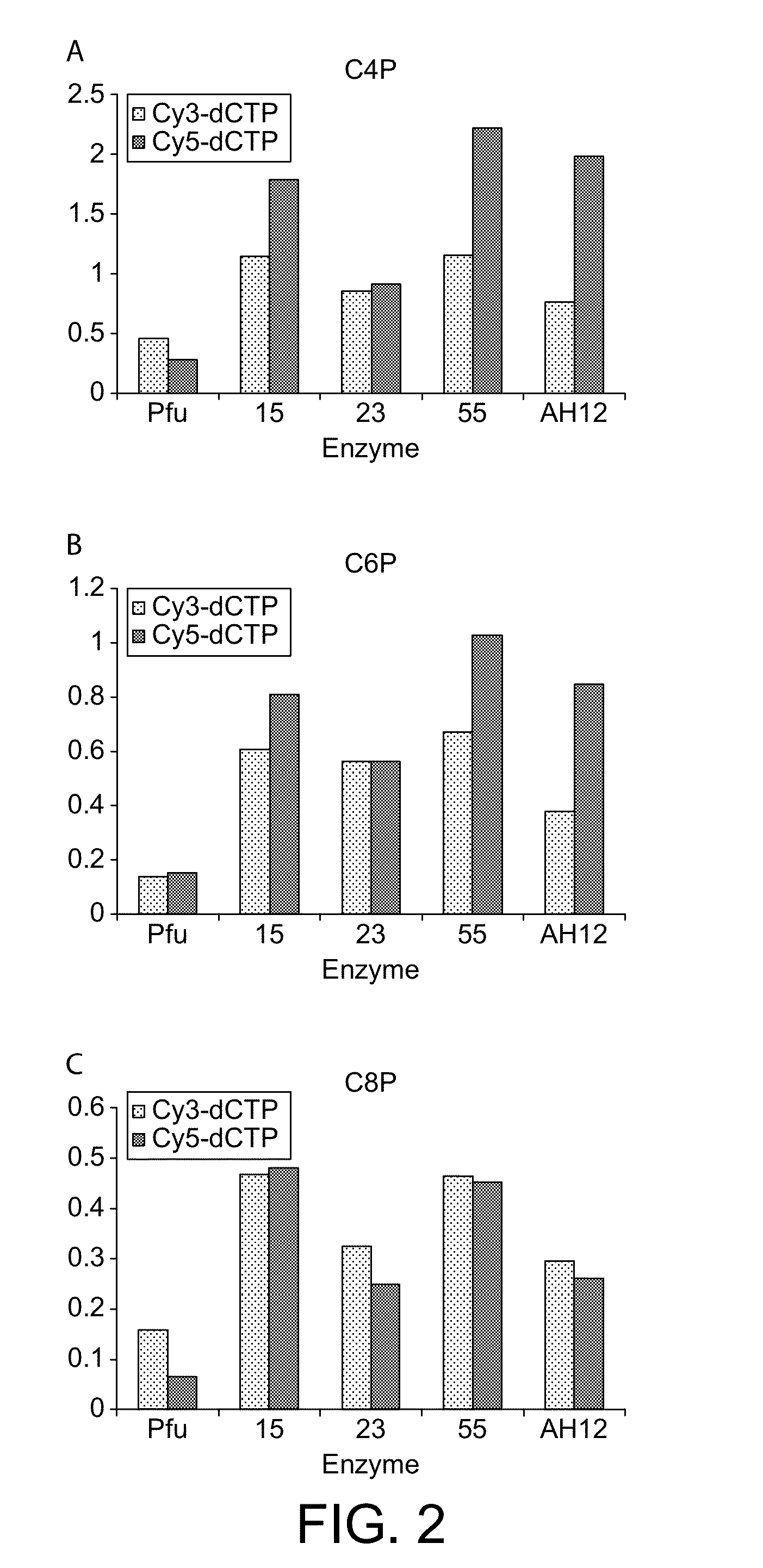
[0214]Modifications to the previously described CSR protocol were required to enable selection for Pfu variants and especially variants of Pfu able to incorporate labelled nucleotide analogues. CSR conditions described previously (Ghadessy et al., 2001), when performed in 1×Pfu buffer (10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-Cl pH 8.75, 2 mM MgSO4, 0.1% Triton X-100, 100 μg / mL BSA; Stratagene Ltd), did not enrich for an active Pfu variant (pASKpfuexo-2; see example 1) over an inactive variant (pASKpfuexo-7; see example 1) when present at a 1:100 ratio. The aqueous phase of the emulsion had to be modified to include primers (1 μM), dNTPs (0.1 mM each), RNase (10 μg / mL), glycerol (10% v / v), formamide (1% v / v), DTT (1 mM) in 1×Pfu buffer. Cells expressing the active Pfu variant were mixed at a ratio of 1:100 with cells expressing the inactive Pfu variant and subjected to PCR with primers 40 and 41 either in solution or in emulsion (CSR) under the modi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com