Plant endosperm specificity promoter and its application
A specific, promoter-based technology, applied in the fields of application, plant genetic improvement, botany equipment and methods, etc., can solve the problem of metabolites that adversely affect biological growth and development, consume bio-energy, and the expression intensity cannot meet the needs of transgenic industrialization, etc. problems, to achieve the effect of increasing added value of science and technology, great application prospects, and improving seed quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, the acquisition of plant endosperm-specific expression promoter (GluC promoter)
[0036] According to the glutelin gene GluC cDNA sequence of rice (GenBank No. AB016501; Mitsukawa et al., 1998, Plant Biotechnol 15: 205-211), the upstream sequence of the GluC gene was searched from GenBank, and primers were designed to amplify the GluC promoter. To facilitate vector construction, two restriction sites (underlined) were sequentially added to each pair of primers.
[0037] Forward primer for GluC promoter is F1: 5'-GGG AAGCTT GTTCAAGATTTTATTTTTGG-3'( Hind III ), the reverse primer is R1: 5'-ACGC GTC GAC AGTTATTCACTTAGTTTCCC-3'( Sal I )
[0038] A small amount of genomic DNA was extracted from rice (wild-type rice Kitaake) leaves by CTAB method, and the GluC promoter sequence was amplified by PCR using it as a template and F1 and R1 as primers. The reaction system is: 1 μl of 10 μM forward and reverse primers, 2 μl of 10×Ex Buffer, 1 μl (10 ng) of ge...
Embodiment 2
[0039] Embodiment 2, plant endosperm-specific expression promoter (GluC promoter) expression vector construction and genetic transformation
[0040] 1. Construction of plant expression vector with GluC promoter fused to GUS gene
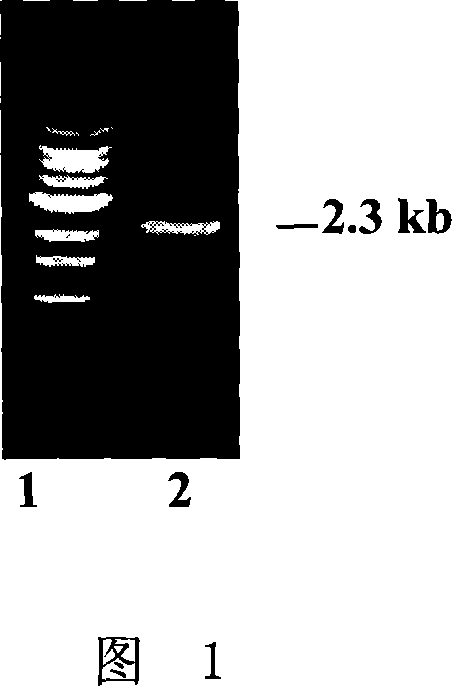
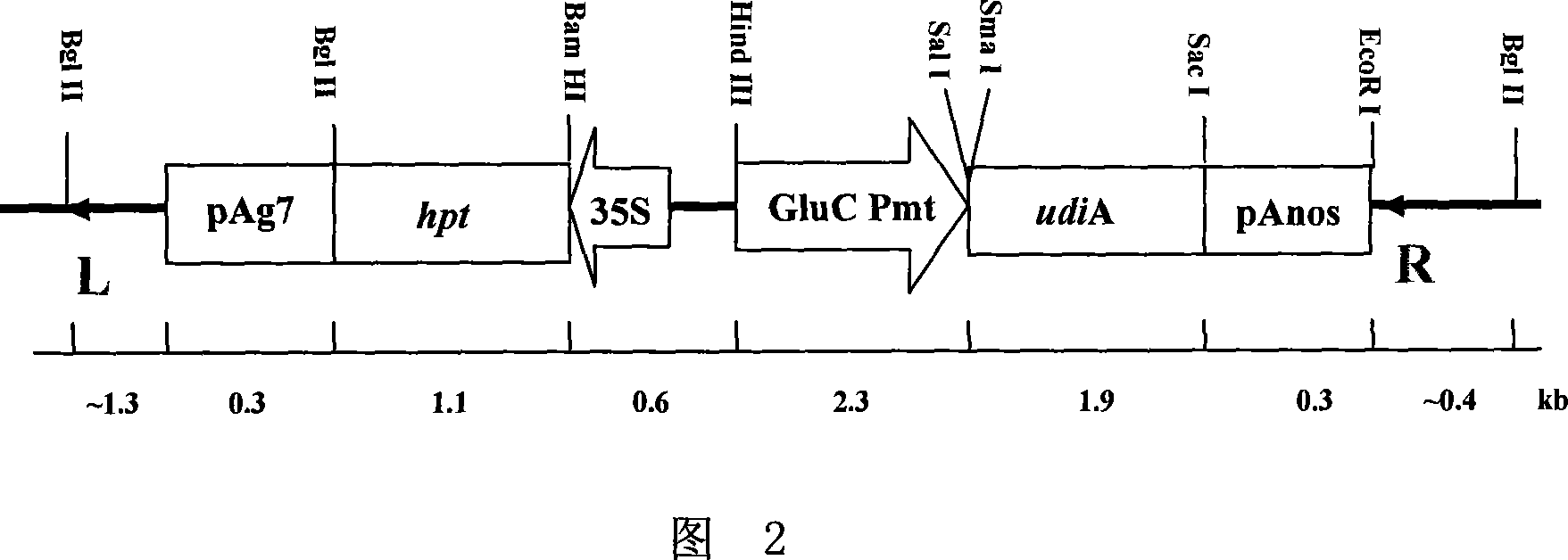
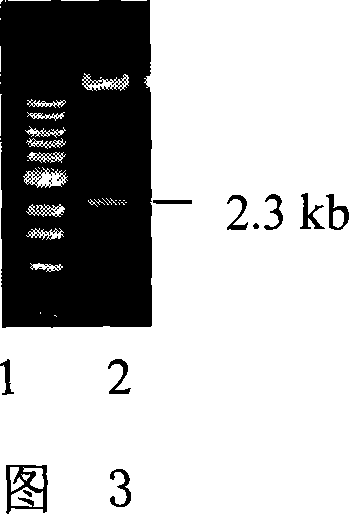
[0041] Digest the pT7-GluC plasmid with Hind III and Sal I, reclaim the 2331bp enzyme-cut fragment containing the GluC promoter, and insert the fragment into pGPTV-35S-HPT (according to the literature Qu and Takaiwa, PlantBiotech J 2: 113-125 between the Hind III and Sal I enzyme recognition sites. On the basis of PCR identification, the obtained recombinant plasmid was identified by double enzyme digestion with Hind III and Sal I, and a fragment containing 4.5kb GluC promoter was obtained; the recombinant plasmid identified by enzyme digestion and identified as containing GluC promoter was named GluC-pGPTV -35S-HPT (the schematic diagram of which is shown in Figure 2). Among them, the double enzyme digestion identification map of GluC-pGPTV-35S-HP...
Embodiment 3
[0049] Example 3, Functional verification of plant endosperm-specific expression promoter (GluC promoter)
[0050] transgenic GluC-pGPTV-35S-HPT rice T 0 Plants were subjected to histochemical staining. Specific steps: Transplant GluC-pGPTV-35S-HPT rice T 0 Substitute plant or wild-type rice Kitaake (control) root, leaf, leaf sheath, stem, cut into small pieces, the seeds of the filling stage 7 days, 12 days, 17 days after flowering were cut longitudinally from the middle with a scalpel, soaked In GUS reaction solution (0.1M NaPO 4 buffer, pH 7.0, 10 mM EDTA, pH 7.0, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1.0 mM X-Gluc, 0.1% Triton X-100), react at 37°C. The results showed that GUS expression was not observed in the roots, leaves, leaf sheaths, and stems of rice transfected with GluC-pGPTV-35S-HPT; the aleurone layer, sub-aleurone layer, and endosperm of the seeds turned blue 7 days after flowering, and the embryos were not stained. Blue; the blue color ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com