Method for separating cotton chloroplast DNA
A chloroplast and cotton technology, applied in the field of plant genetic engineering, can solve the problems of expensive, time-consuming, low yield, etc., and achieve the effect of saving experiment cost, low price and high quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] The experimental material in this example is the upland cotton standard line Tm-1 (Lin et al., 2003). Take 50g of young cotton leaves planted in the field or in a tray, wash them with distilled water, dry them slightly, put them in a fresh-keeping bag and starve them overnight in a dark place, or store them at -70°C for later use.
[0059] 1. Isolation of intact chloroplasts
[0060] (1) Take the newly expanded young leaves of cotton plants in the field as materials for separating cotton chloroplasts, place the leaves in a refrigerator (4°C) for 24 hours in the dark, and then transfer them to an ultra-low temperature (-70)°C refrigerator for storage (or directly use About 50 grams of leaves were weighed for separation of chloroplasts. Cool the juicer with ice cubes in advance, then add 4 times the volume of sample pre-cooled buffer A at 4°C to the juicer, quickly add the sample to the container, start the juicer, homogenize for about 1min, and then use Filter the homo...
Embodiment 2
[0100] Gel electrophoresis and UV spectrophotometer to detect the quality of cpDNA:
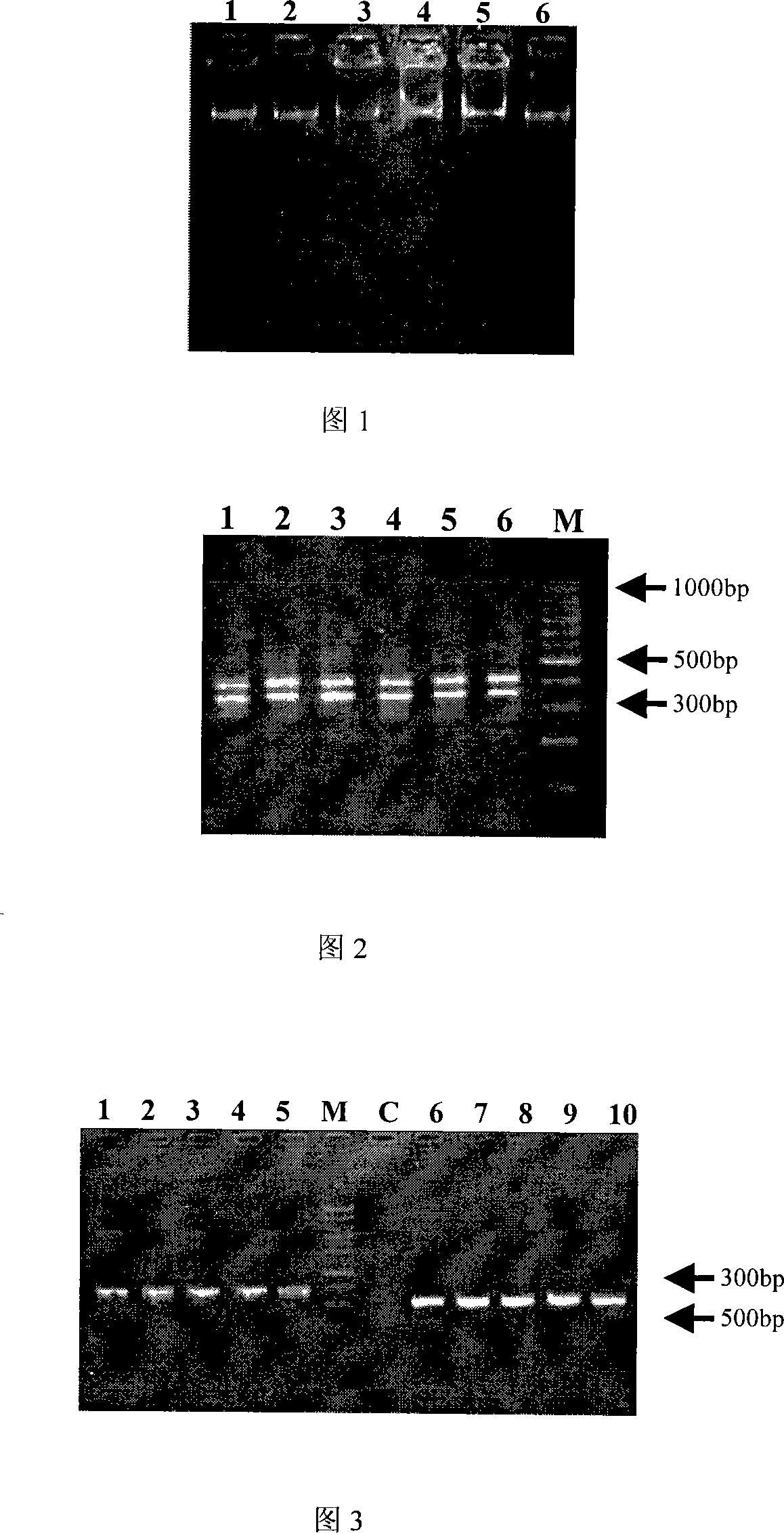
[0101] 5 μL of extracted chloroplast DNA was electrophoresed on 0.8% agarose gel with 90V×60mA for 30min, and placed in a BIO-RAD gel imaging system to observe and image the results. After 4 μL of cpDNA was diluted 50 times, D260nm and D280nm were measured on a Beckman-DU 800 UV spectrophotometer.
[0102] It can be seen from Figure 1 that the size of the extracted cpDNA fragment is about 20,000 bp, and the band pattern is uniform, indicating that the purity of the cpDNA is high, and there is no obvious tailing and dispersion phenomenon, indicating that the integrity of the extracted DNA is relatively good.
[0103] The results of ultraviolet spectrophotometry can also be seen that the D of most samples 260 / D 280 Between 1.6-1.8, it shows that the purity of cpDNA is higher (Table 1).
[0104] Table 1 Cotton cpDNA D 260 and D 280 The measurement results
[0105] the batch
...
Embodiment 3
[0111] PCR detection of DNA extraction and purification effect:
[0112] The promoter and terminator primers of the psbA gene were designed as follows:
[0113] Promoter primer: forward primer: 5'-CCCGGGCAACCCATTTTTAGTATC-3'; reverse primer:
[0114] 5'-TTAATCATCAGGGACTCCCAAGCG-3'.
[0115] Terminator primer: forward primer 5′AGACTTTGGTTTTAGTGTATACGAG 3′;
[0116] Reverse primer 5'-TGCCTTGATCCACTTGGCTACA-3'.
[0117] The expected amplified fragments are 310, 380bp psbA gene promoter and terminator fragments respectively.
[0118] 20ul system for PCR amplification procedure:
[0119] Taq polymerase 1mol / L, 25mmol / L MgCl2 1.5μL, 10mmol / L dNTP 0.5μL, 10×Buffer 2.5μL, 10μmol / L forward and reverse primers 0.25μL each, template DNA 40ng, total volume 25μL. PCR reaction program: 94°C pre-denaturation for 5 min, followed by 94°C denaturation for 1 min, 58°C annealing for 30 s, 72°C ligation for 1 min cycle 38 times, and finally 72°C extension for 5 min. The PCR reaction was car...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com