Mononucleotide polymorphism (SNP) high-pass typing method based on magnetic nano particle polymerase chain reaction (PCR)
A magnetic nanoparticle and single nucleotide polymorphism technology, applied in the field of single nucleotide polymorphism typing, can solve the problems of high cost, inability to give sequence information, high operator requirements, etc., and achieve positive mismatch The effect of high number ratio, intuitive typing results and high application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
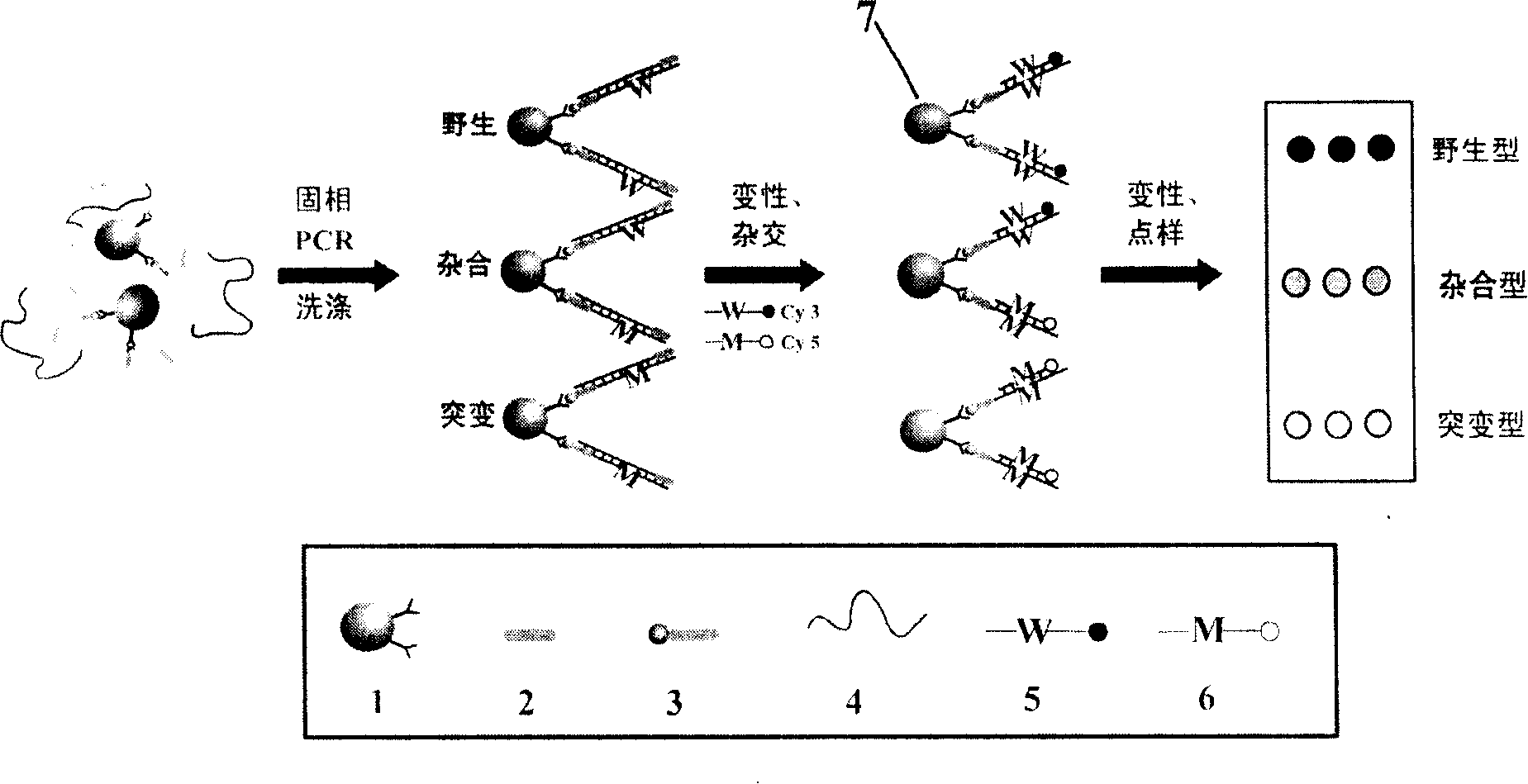
[0022] The invention relates to a high-throughput SNPs detection method. The details are as follows: 1. Polymerase chain reaction based on magnetic nanoparticles: biotin-labeled primers are immobilized on avidin-coated magnetic nanoparticles, and carried out in a system containing free primers, dNTPs, polymerase and template DNA The PCR reaction directly amplifies the target fragment (MNPs-DNA) containing the detected SNP site on the surface of the magnetic nanoparticle. 2. Denaturation and quenching, the double-stranded PCR product on the surface of the magnetic particle becomes single-stranded (MNPs-ssDNA). 3. Hybridization: Design a pair of allele-specific detection probes (wild-type Cy3 marker, mutant Cy5 marker) according to the sequence at the gene site to be tested, and the ssDNA immobilized on the surface of the particle and the designed probe are Hybridization at a specific temperature. 4. Fully wash to remove the free target sequence, denature, spot the denatured f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com