Mass spectrometry reagent kit and method for rapid tuberculosis diagnosis
A kit and mass spectrometry technology, applied in the field of protein detection, can solve the problems of high cost, low positive rate and long time-consuming of tuberculosis polymerase chain reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Example 1 The distinction between normal and tuberculosis and the preparation of a mass spectrometry kit
[0087] (1) Experimental method
[0088] 1. Materials
[0089] Specimen source: Standardized quality control serum (plasma) prepared by mass spectrometry. The definition meets the following criteria. The blood donors are half male and half male, and the blood type is type 0; the age is 18-30 years old; ethnicity, Han. The biochemical indicators were normal, including: total cholesterol, triglycerides, fasting blood sugar, hepatitis B surface antigen, liver function test, kidney function test; no family history of genetic diseases; no history of major infectious diseases. Women without pregnancy, men with no history of smoking. Fresh blood was drawn from 10 healthy subjects with type 0 blood (half male and half female), and centrifuged at 10,000 rpm for 5 minutes at 4°C immediately after the blood coagulated; the samples were stored at -80°C.
[0090] Take 10 μl o...
Embodiment 2
[0105] The double-blind test of embodiment 2 kit
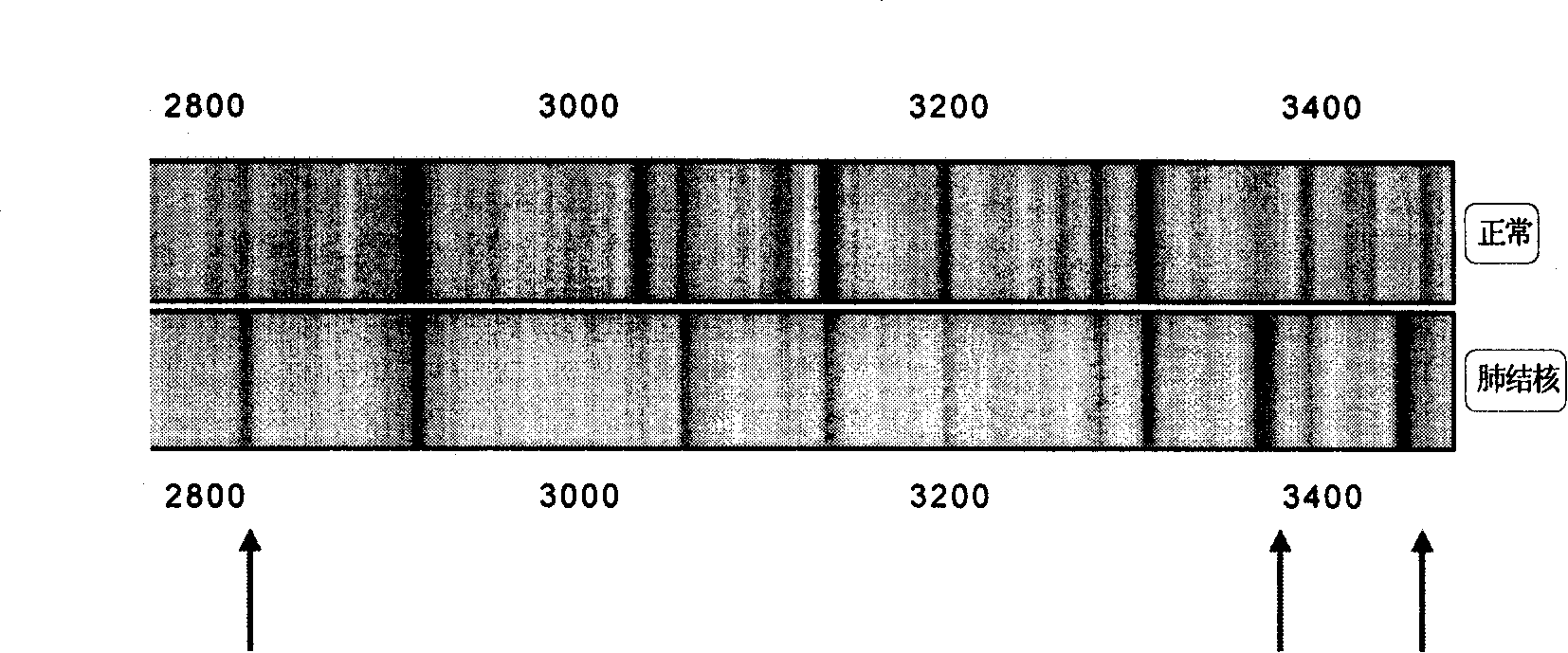
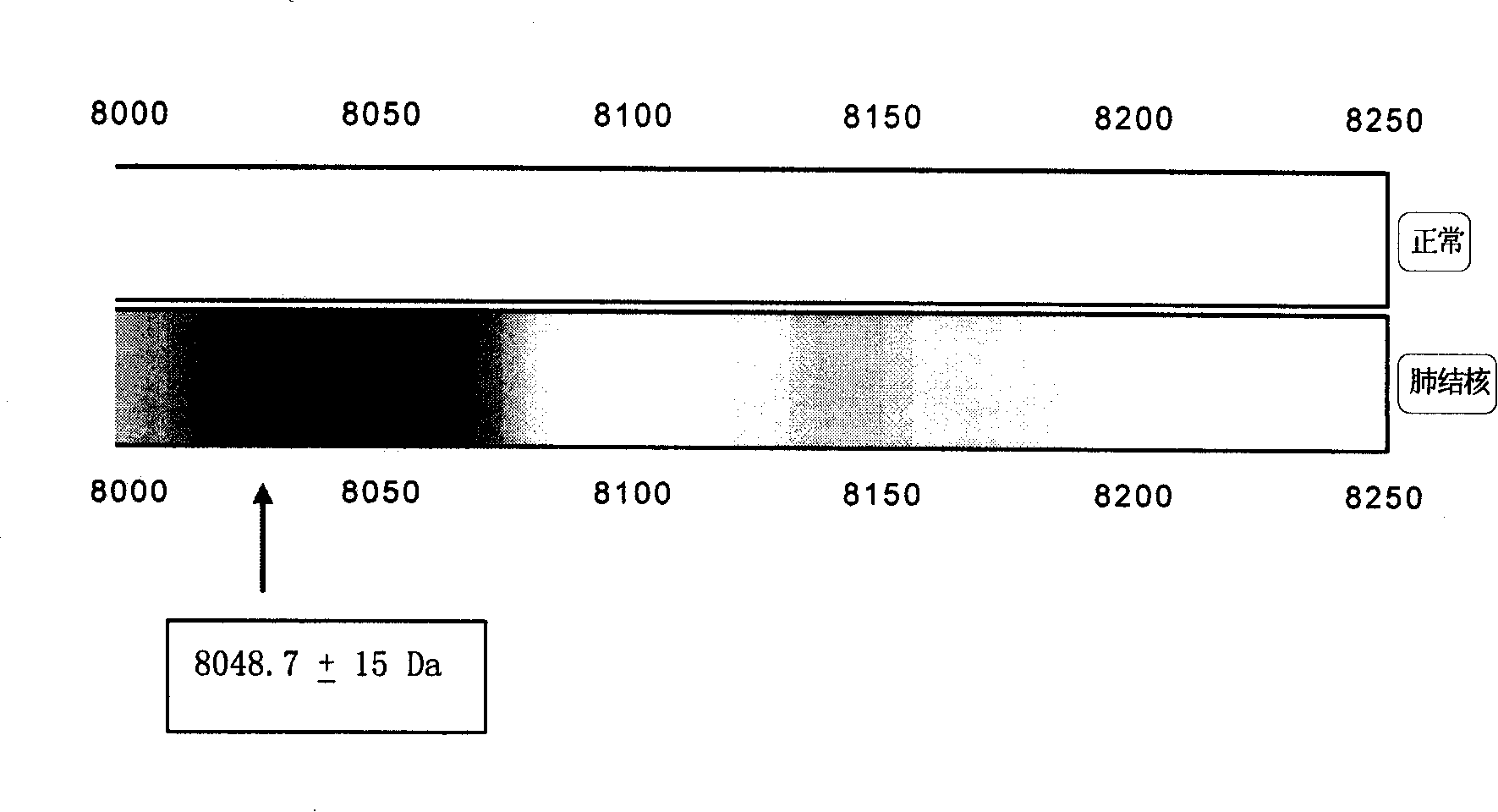
[0106] Several characteristic protein peaks 2873.3±15Da, 3379.7±15Da, 3449.3±15Da, 5808.4±15Da, 7568.9±15Da, 8048.7±15Da, 11439.2±15Da, 11526.7±15Da, 11683.0±15Da, 15114.9 were screened out from Example 1 ±15Da, 15312.8±15Da, double-blind test of 100 cases of normal and 100 cases of tuberculosis:
[0107] double blind test
Tuberculosis (100)
normal(100)
tuberculosis
100
0
normal
0
100
[0108] Sensitivity 100% Specificity 100%
[0109] The experimental results using C8 and C18 hydrophobic matrix beads are consistent with the experimental results of the above-mentioned WCX anion matrix beads.
[0110] Experimental results
[0111] The kit of the method can distinguish between normal people and tuberculosis, with 100% sensitivity and 100% specificity.
Embodiment 3
[0112] Example 3 Sorting identification of a group of characteristic peaks of tuberculosis protein fingerprint
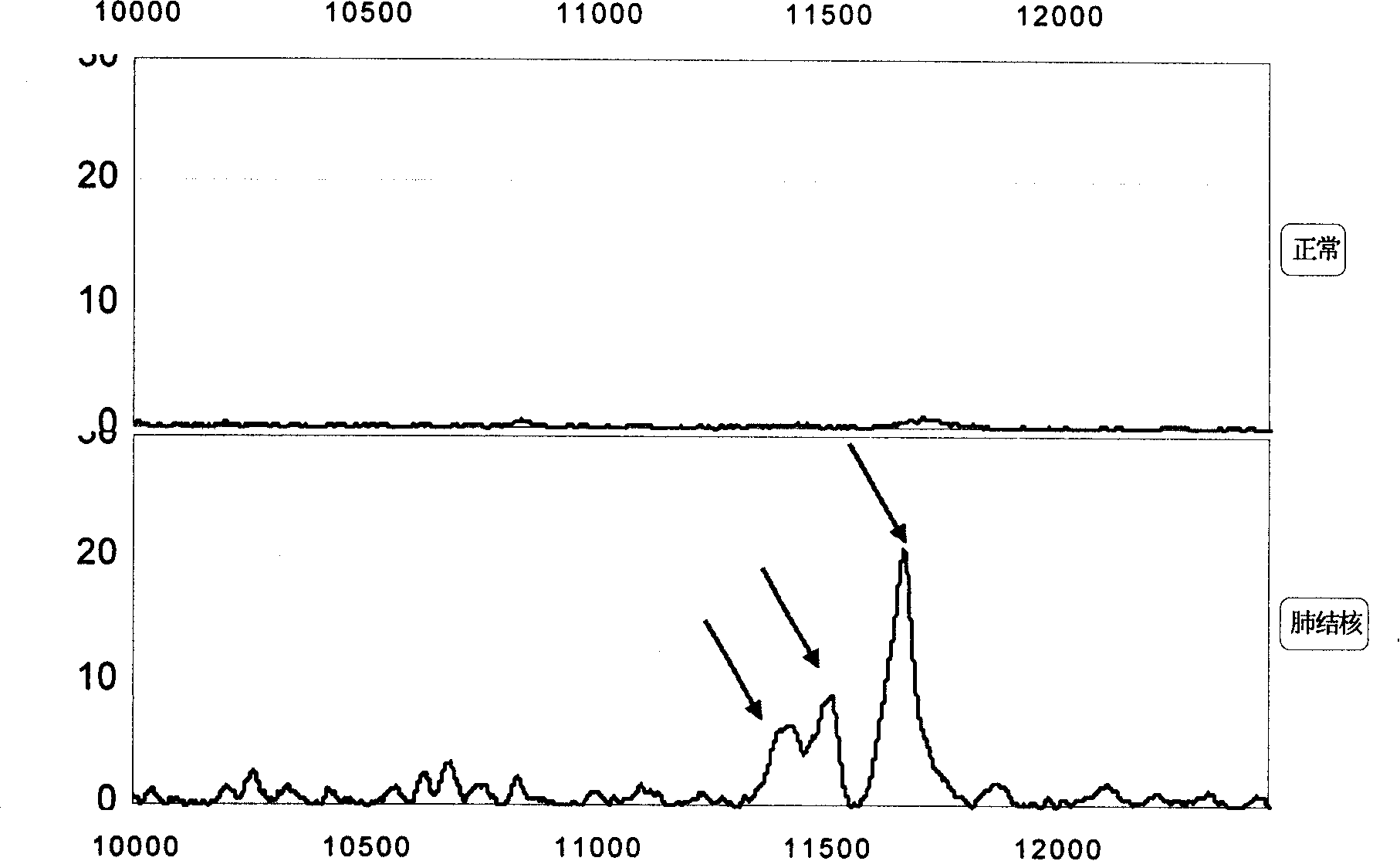
[0113] A group of 11439.2±15Da, 11526.7±15Da, 11683.0±15Da biomarkers ( image 3 ) by multiple-stage mass spectrometry (MS / MS), post-source fragmentation (PSD) and protein ladder sequencing (protein ladder sequencing) for sequencing and other methods. By breaking molecules into pieces, protein ladders can be generated. This gradient is then analyzed by mass spectrometry. The 11439.2±15Da, 11526.7±15Da, and 11683.0±15Da biomarkers were identified as variant serum amyloid A. Its chemical structure is (104 amino acids arranged from N-terminal to C-terminal):
[0114] Chemical structures of 11683 ± 15 Da biomarkers:
[0115] N-terminal
[0116] RSFFSFLGEAFDGARDMWRAYSDMREANYIGSDKYFHARGNYDAAKRGPG
[0117] GVWAAEAISDARENIQRFFGHGAEDSLADQAANEWGRSGKDPNHFRPAGL
[0118] PEKY
[0119] C-terminal.
[0120] Chemical structure of the 11527±15 Da biomarker:
[0121] N-termi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com