Use of acid stable protease in animal feed
A technology of animal feed and protease, which is applied in the field of protease treatment of plant protein to achieve good acid-stability and improved performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 7
[0118] Examples of animal feed compositions are shown in Example 7.
[0119] Table B Value ranges for energy, protein and minerals in animal foods
[0120] Nutrients
poultry
piglet / pig / big
sow
fish
R.1
R.2
R.3
R.4
R.5
lowest-highest
lowest-highest
lowest-most
high
Metabolizable
energy,
MJ / kg
12.1-13.4
12.9-13.5
14-25
10-30
11-28
11-26
12-25
crude protein,
g / kg
124-280
120-240
300-480
50-800
75-700
100-600
110-500
120-490
Calcium, g / kg
8-40
5-9
10-15
0.1-200
0.5-150
1-100
4-50
Phosphorus, g / kg
2.1-6.0
1.5-5.5
3-12
0.1-200
0.5-150
1-100
1-50
1-25
Acid, g / kg
3.2-5.5
-
12-16...
Embodiment 1
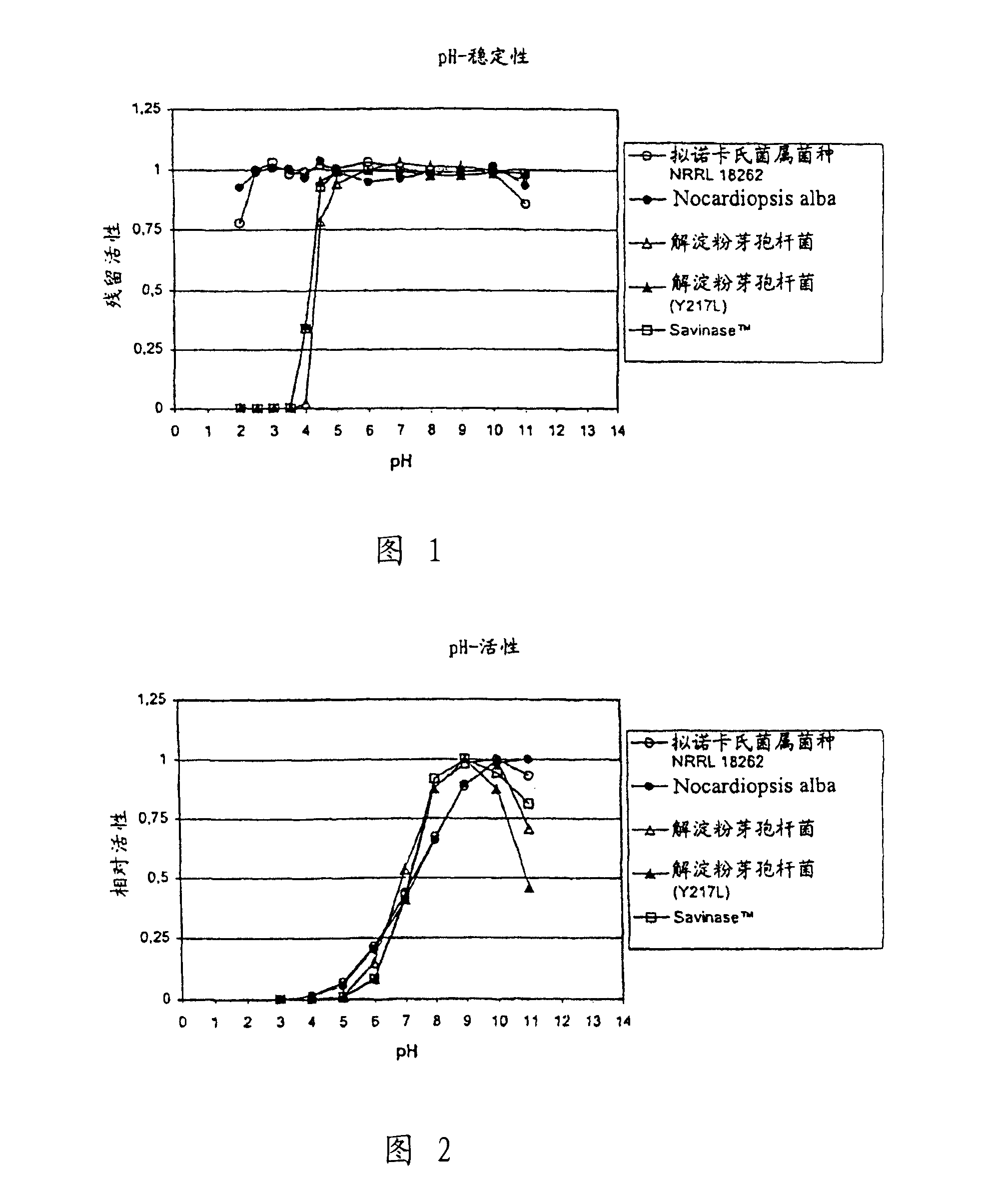
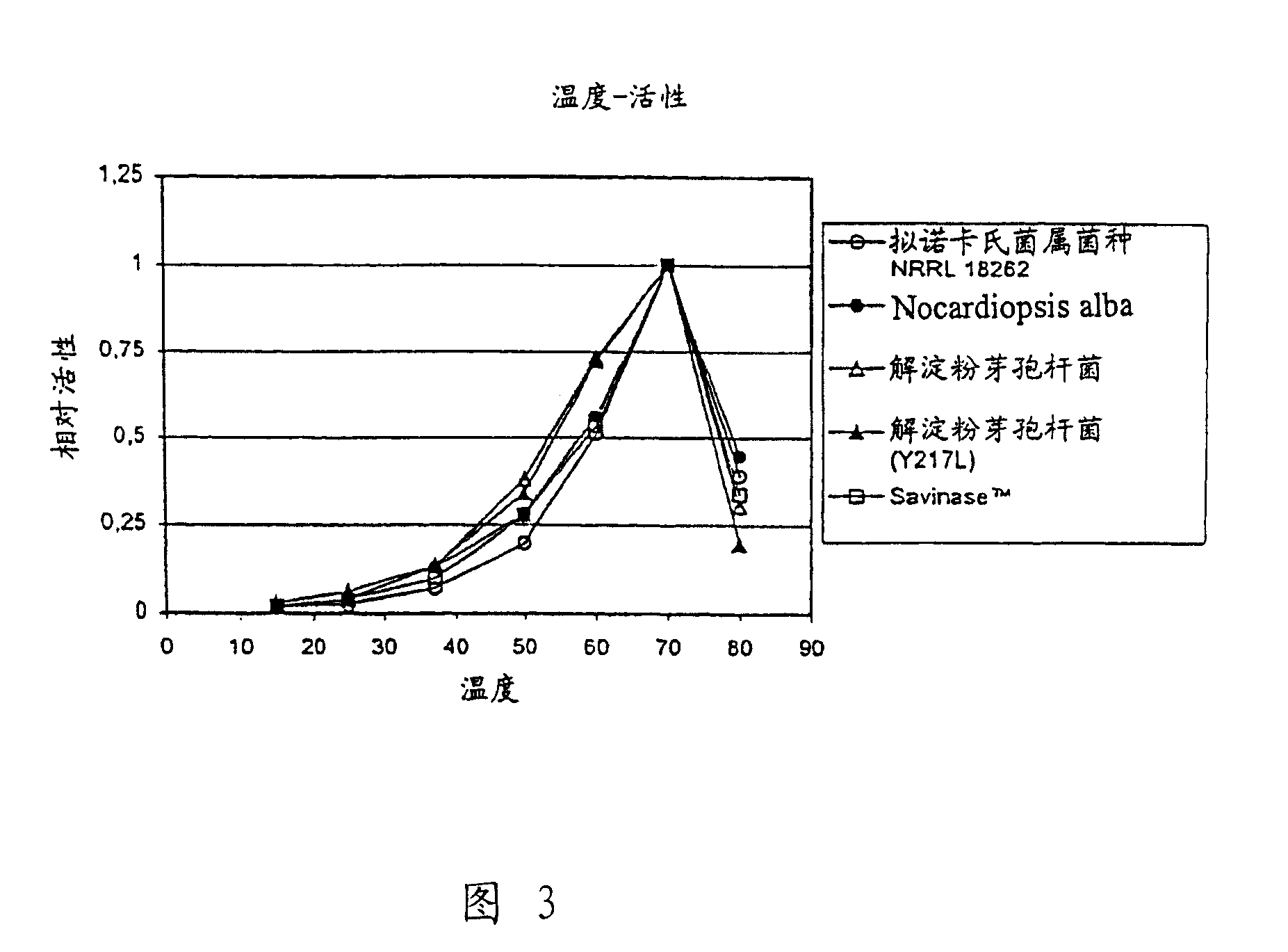
[0133] Screening for acid-stable proteases
[0134] The stability of various proteases at pH 3 was analyzed with the aim of identifying proteases with the necessary stability to flow through the acidic stomach of monogastric animals.
[0135] By conventional chromatography, such as ion exchange chromatography, hydrophobic interaction chromatography and size exclusion chromatography (see for example Protein Purification, Principles, High Resolution Methods, and Applications, Jan-Christer Janson, edited by Lars Ryden, VCH Publishers, 1989 ) to purify protease.
[0136] Protease activity was determined as follows: protease was incubated with 1.67% Hammarsten casein at 25°C, pH 9.5 for 30 minutes, then TCA (trichloroacetic acid) was added to a final concentration of 2% (w / w), and the mixture was filtered to The precipitate was removed and the filtrate was analyzed for free primary amino groups (detected in a colorimetric assay based on OPA (o-phthalaldehyde) by measuring the ab...
Embodiment 2
[0143] Preparation, Identification and Comparative Study of Protease from Nocardiopsis
[0144] fermentation
[0145] Nocardiopsis alba was inoculated from tryptone yeast agar plates into flasks each containing 100 ml of HG-23 medium with the following composition: oat flour 45 g / l, yeast extract 2 g / l, disodium hydrogen phosphate 12 g / l 1. Potassium dihydrogen phosphate 6g / l, Pluronic PE 6100 0.2ml / l, dissolved in distilled water. The strain was fermented at 37°C for 9 days.
[0146] purification
[0147] The culture was centrifuged at 10000 x g for 30 minutes in a 1 liter beaker. The supernatants were pooled and further clarified by filtration through Seitz K-250 depth filter plates. The clarified filtrate was concentrated by ultrafiltration over a polyethersulfone cartridge (Filtron) with a molecular weight cut-off of 3 kDa. The concentrated enzyme was transferred to 50 mM H on a G25 Sephadex column (Amersham Pharmacia Biotech). 3 BO 3 , 5 mM 3,3′-dimethylgluta...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com