Method for detecting DNA, RNA and ultramicro-amount protein
A protein, ultra-trace technology, applied in the field of DNA detection, can solve the problems of false positives, low sensitivity and high requirements for experimental conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
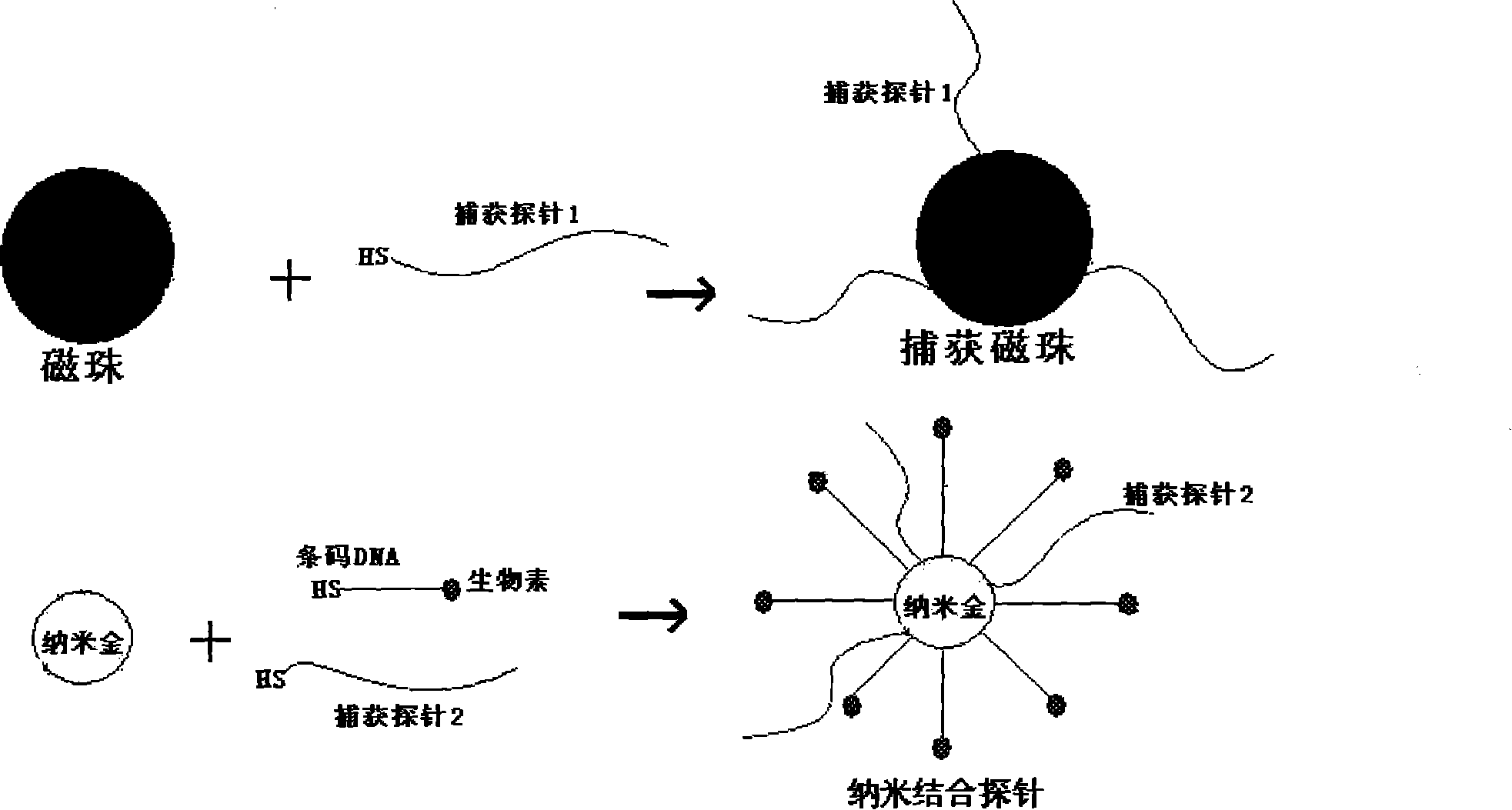
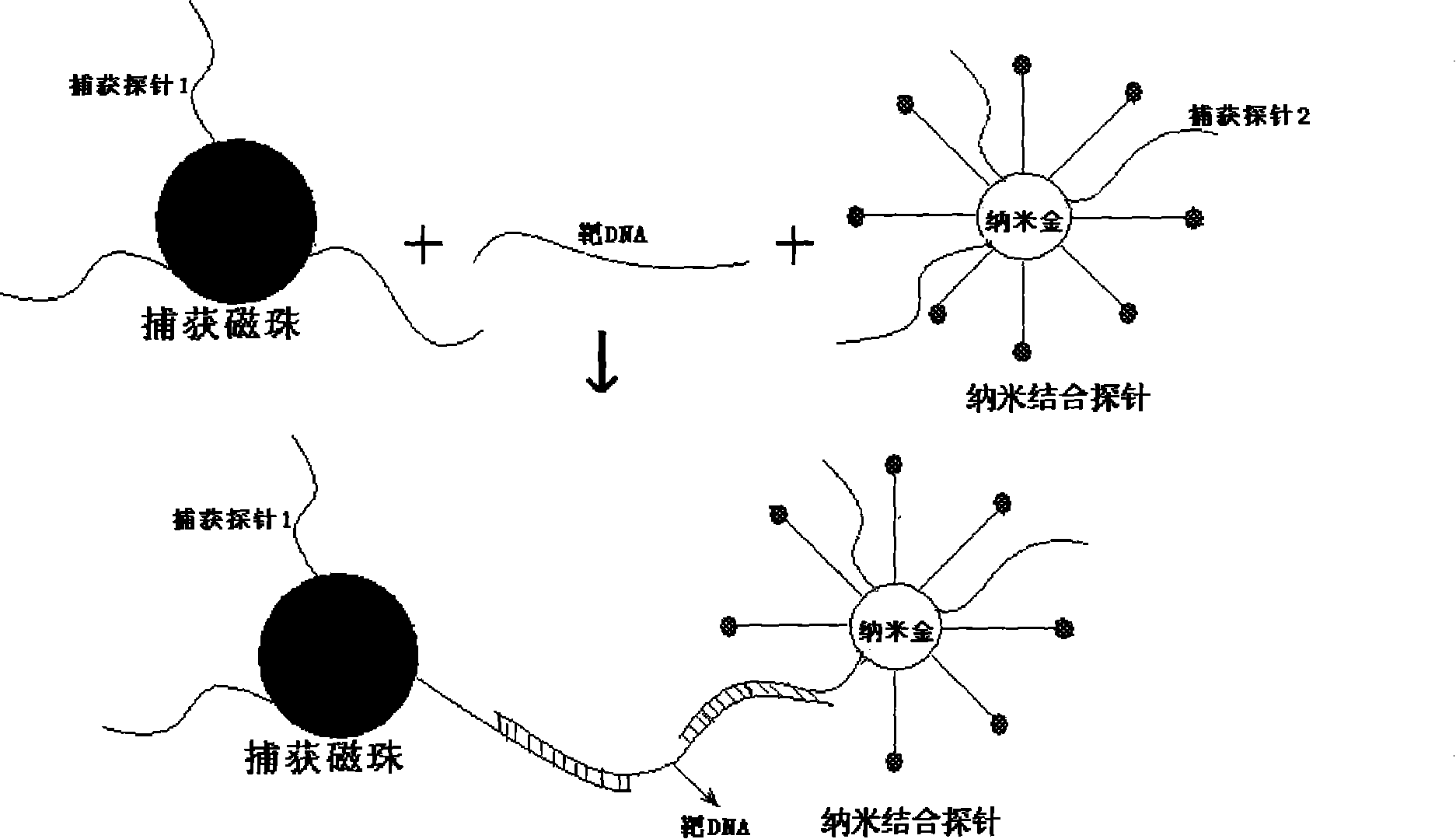
[0014] 1. Preparation of magnetic bead capture probe: magnetic beads with free-NH3 on the surface, and capture probe 1 with -SH at the end (3 with -SH at the end, PEG and 10 consecutive A in the middle, As a spacer bridge sequence, the 5-terminus is a 18-25bp base sequence complementary to the 3-terminus of the detection target DNA) Use glutaraldehyde to connect the amino group and the sulfhydryl group, so that the capture probe 1 is labeled on the surface of the magnetic bead to make a magnetic bead capture probe ( figure 1 ).
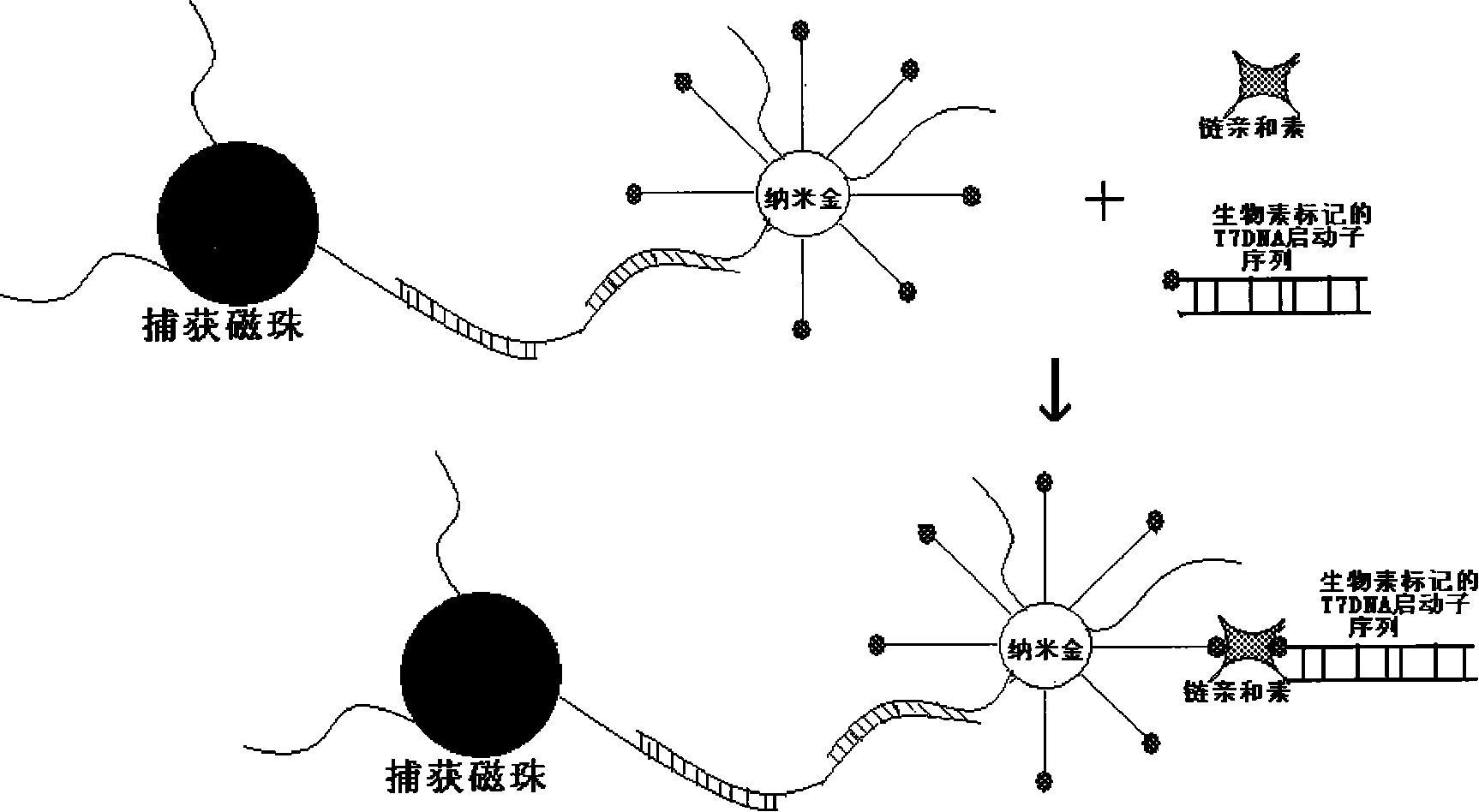
[0015] 2. Preparation of nano-binding probes: gold atoms of nano-gold particles form covalent bonds with capture probe 2 and barcode DNA (barcode DNA / capture probe 2 is 100:1) with -SH at the 5-terminus in aqueous solution , firmly bound to the surface of gold nanoparticles, gradually adding NaCl to a final concentration of 0.3M, so that the capture probe 2 and the barcode DNA stretch on the surface of gold nanoparticles (capture probe 2 has -SH at i...
Embodiment 1
[0022] Embodiment 1: the detection of DNA
[0023] 1. Primer design: Take the detection of the BamHI-W fragment DNA of EBV as an example (NCBI genbankaccession No: M15973).
[0024] Detect the sequence of BamHI-W fragment DNA:
[0025] 2091 CCATCCCTGA AGACCCAGCGGCCATTCTCTCTGGTAACGAGCAGAGAAGA
[0026] AGTAGAGGCC CGCGGCCATT GGGCCCAGAT TGAGAGACCA GTCCAGGGGC CCGAGGTTGG
[0027] AGCCAGCGGG CACCCGAGGT CC 2222.
[0028] (1), capture probe 1:5- GGG CTT GGC CGG GTC TAA -PEG-AAAAAAAAAA-SH-(C6)-3. (The underlined part is combined with EBV BamHI-W fragment DNA).
[0029] (2), capture probe 2: 5-SH-(C6)-AAAAAAAAAA-PEG- GGA GGG ACC GGG TGC TGG-3 (The underlined part is combined with EBV BamHI-W fragment DNA).
[0030] (3) Barcode DNA: 5-SH-(C6)-AAA AAA AAA A-PEG-GGA TTC TCA ACTCGT AGC T-Biotin-3.
[0031] (4) Positive detection of target DNA: Use EBV BamHI-W fragment DNA as a template to synthesize a positive control primer, 5-CCA GCA CCC GGT CCC TCC AAA AAA AAA TTA GAC CCGGCC...
Embodiment 2
[0061] Embodiment 2: Detection of RNA
[0062] 1. Primer design: Take the detection of HCV RNA as an example (NCBI accession No: EU482888, detection corresponding sequence nt 1-216).
[0063] (1), capture probe 1:5- AAC TAC TGT CTT CAC GCA GAA AGC -PEG-AAAAAAAAAA-SH-(C6)-3. (The underlined part binds to HCV 5-UTR nt 1-18 RNA).
[0064] (2), capture probe 2: 5-SH-(C6)-AAAAAAAAAA-PEG- CCC AAC ACT ACT CGG CTA G-3 (The underlined part binds to HCV 5-UTR nt 198-216 RNA).
[0065] (3) Barcode DNA: 5-SH-(C6)-AAAAAAAAAA-PEG-GGA TTC TCA ACT CGTAGC T-Biotin-3.
[0066] (4) Positive detection of target RNA: WHO HCV standard.
[0067] 2. Preparation of magnetic bead capture probe:
[0068] (1) Take 1ml of magnetic beads with a diameter of 1 μm and free-NH3 on the surface, add 4ml of PBS (0.01M, pH 7.4,) and shake well, absorb the magnetic beads with a magnet, and remove the supernatant; repeat three times, remove the supernatant.
[0069] (2) Prepare fresh 5% glutaraldehyde solu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com