Visual detection method for PCR (polymerase chain reaction) amplification and endpoint
A chain reaction and detection method technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as cross-contamination, endanger the health of operators, and restrict promotion and application, achieve the image of test results, and avoid false positives. Positive questions, the effect of simplifying the testing process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Design of primers for the amplification detection of human pathogenic Mycobacterium tuberculosis specific gene IS6110 The 812-1045 nucleic acid sequence of human pathogenic Mycobacterium tuberculosis IS6110 was screened by reviewing the literature and analyzing with BLAST software, and the two fragments Site (the two sites: 812–831bp and 1029–1045bp respectively) designed and synthesized PCR primers to obtain the following primers; the primer design was completed by PCR-specific primer design software. Forward primer F-MYC-IS6110AGACCTCACCTATGTGTCGA Reverse primer R-MYC-IS6110TCGCTGAACCGGATCGA. Reaction system (total reaction volume is 30μL): 100ng nucleic acid template, 10uM F-MYC-IS6110, 10μM R-MYC-IS6110, 0.2μM dNTP, 0.2mM visible light dye, 1X PCR Buffer, 5U Taq DNA Polymerase, supplemented with ddH 2 0 to 30 μL.
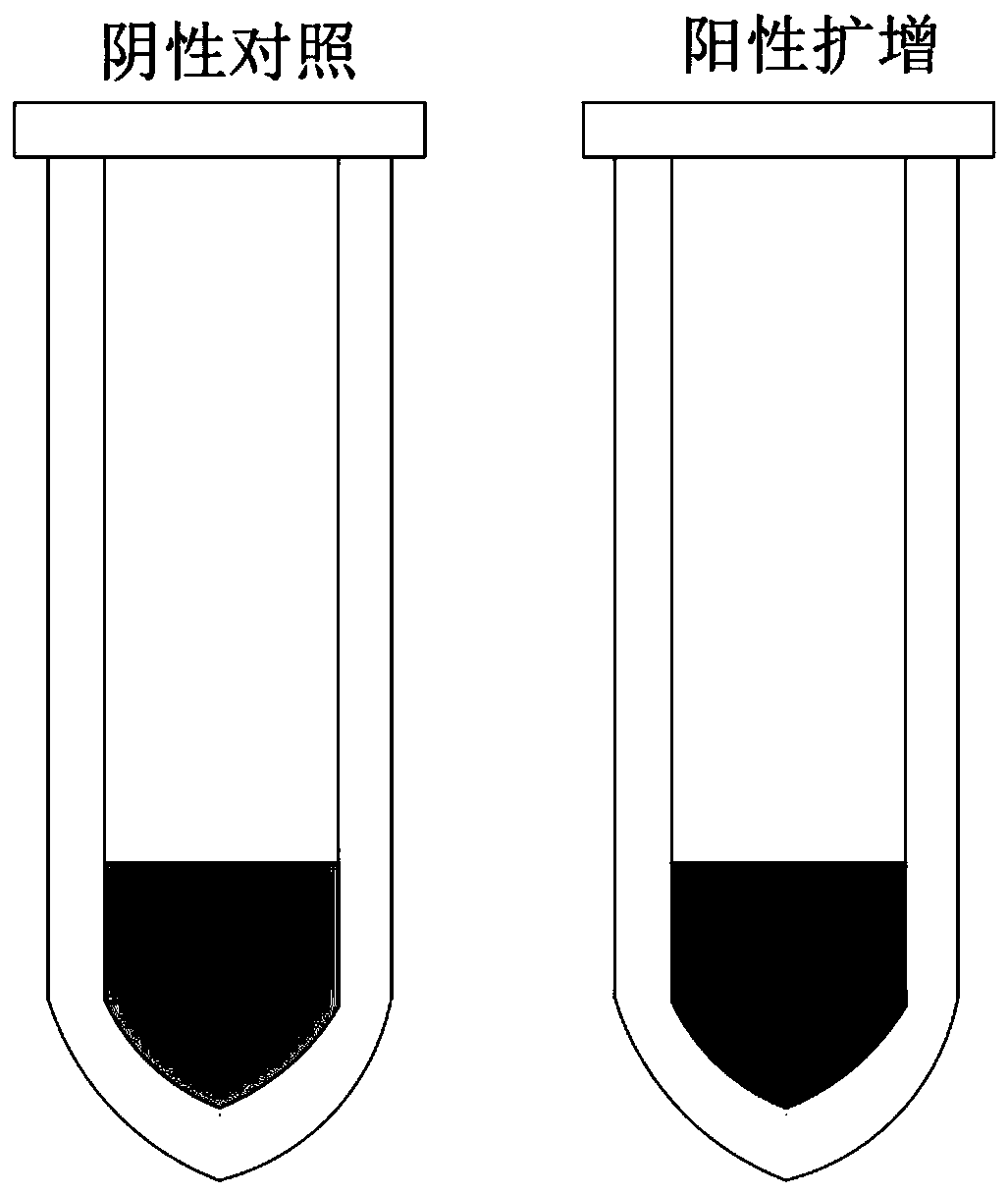
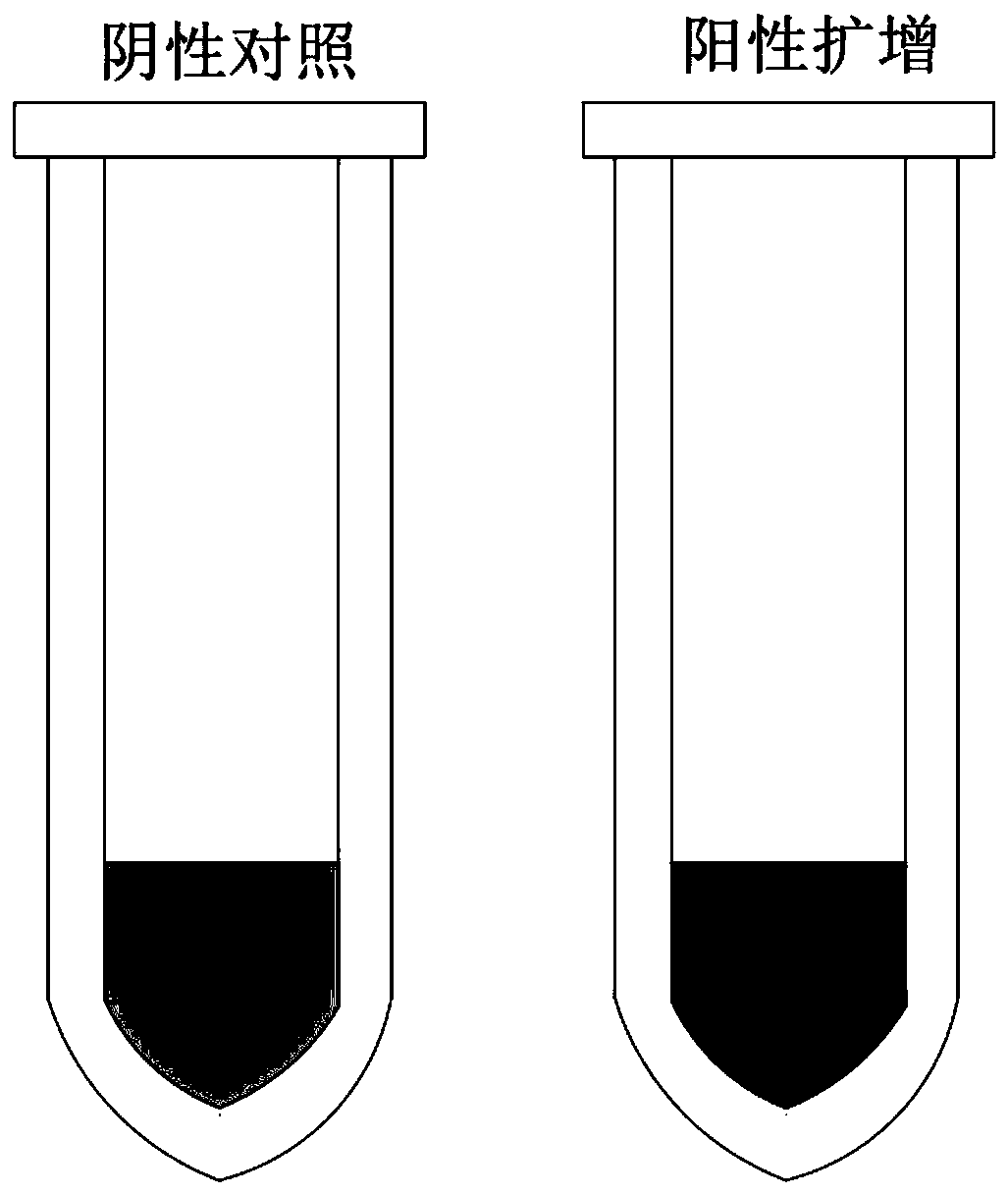
[0026] Amplification reaction program: 95°C for half a minute, 50°C for half a minute, 72°C for half a minute, 30 cycles. The results judge that after ...
Embodiment 2
[0028] Design of primers for the amplification detection of Shigella specific gene ipaH in food The 232-437 nucleic acid sequence of Shigella specific gene ipaH was screened out by consulting the literature and using BLAST software analysis, targeting two sites of this fragment (These two sites are located at 233-254bp and 420-438bp respectively) PCR primers were designed and synthesized to obtain the following primers; the primer design was completed by PCR-specific primer design software. Forward primer F-ipaHCATGGCTGGAAAAACTCAGTGC Reverse primer R-ipaHGAGGCGGAACATTTCCCTG. Reaction system (total reaction volume is 30μL): 100ng nucleic acid template, 10uMF-ipaH, 10μM R-ipaH, 0.2μM dNTP, 0.1-0.3mM visible light dye, 1X PCR Buffer, 5U Taq DNAPolymerase, supplemented with ddH 2 0 to 30 μL.
[0029] Amplification reaction program: 95°C for half a minute, 52°C for half a minute, 72°C for half a minute, 35 cycles. The results judge that after the reaction is over, the reaction sy...
Embodiment 3
[0031] Amplification and detection primer design of Macrobrachium rosenbergii nodavirus RNA-2 Through literature review and BLAST software analysis, the specific gene fragment RNA-2 of Macrobrachium rosenbergii Nodavirus was screened out, targeting two sites of the fragment (these two The two sites are respectively: 1976-1994bp and 2198-2216bp) PCR primers were designed and synthesized to obtain the following primers; the primer design was completed by PCR-specific primer design software. Forward primer F-RNA-2CCAATATGAACCGGGAGTG Reverse primer R-RNA-2GGGTTCAACCTTGAGTTCC. Reaction system (total reaction volume is 30μL): 100ng nucleic acid template, 10uM F-RNA-2, 10μM R-RNA-2, 0.2μM dNTP, 0.1-0.3mM visible light dye, 1X PCR Buffer, 5U Taq DNAPolymerase, supplemented with ddH 2 0 to 30 μL.
[0032] Amplification reaction process: 95°C for half a minute, 53°C for half a minute, 72°C for half a minute, 40 cycles. The results judge that after the reaction is over, the reaction sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com