Chikungunya virus testing method
A chikungunya virus and detection method technology, applied in the field of molecular biology detection, to achieve important social and economic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1, design and synthesis of primers and probes
[0030] The SeqMan program in the Lasergene software package was used to analyze the nucleotide sequences of 37 strains of Chikungunya virus obtained from Genbank, select non-structural protein genes with highly conserved nucleotides, and use Primer Express2.0 software to design specific primers and The probe design, primers and probe sequences are listed in Table 1.
[0031] Table 1
[0032]
[0033] The location of the primers and probes on the Chikungunya virus genome was based on the strain sequence with the GenBank sequence number AF490259.
Embodiment 2
[0034] Example 2. Preparation of positive control RNA template and extraction of viral RNA for specificity assessment
[0035] According to the sequence of Chikungunya virus (GenBank sequence number: AF490259), synthesize a DNA fragment with a size of 120bp including the fragment sequence detected by fluorescent RT-PCR, clone it on the T vector (TOPO TA cloning kit), and use plasmid DNA extraction reagent Purify the plasmid DNA with the cassette, and after sequencing confirms that the inserted fragment is correct, reverse transcription in vitro: first use Not I to digest the plasmid to linearize it to exclude downstream RNA products, then perform in vitro transcription with MAXIscript T7 Kit to generate the target RNA, and then use Turbo DNase DNase treatment was performed to remove the untranscribed plasmid DNA template, and finally ethanol precipitation was used to purify the RNA product, which was the positive control RNA template and stored in a -70°C ultra-low temperature ...
Embodiment 3
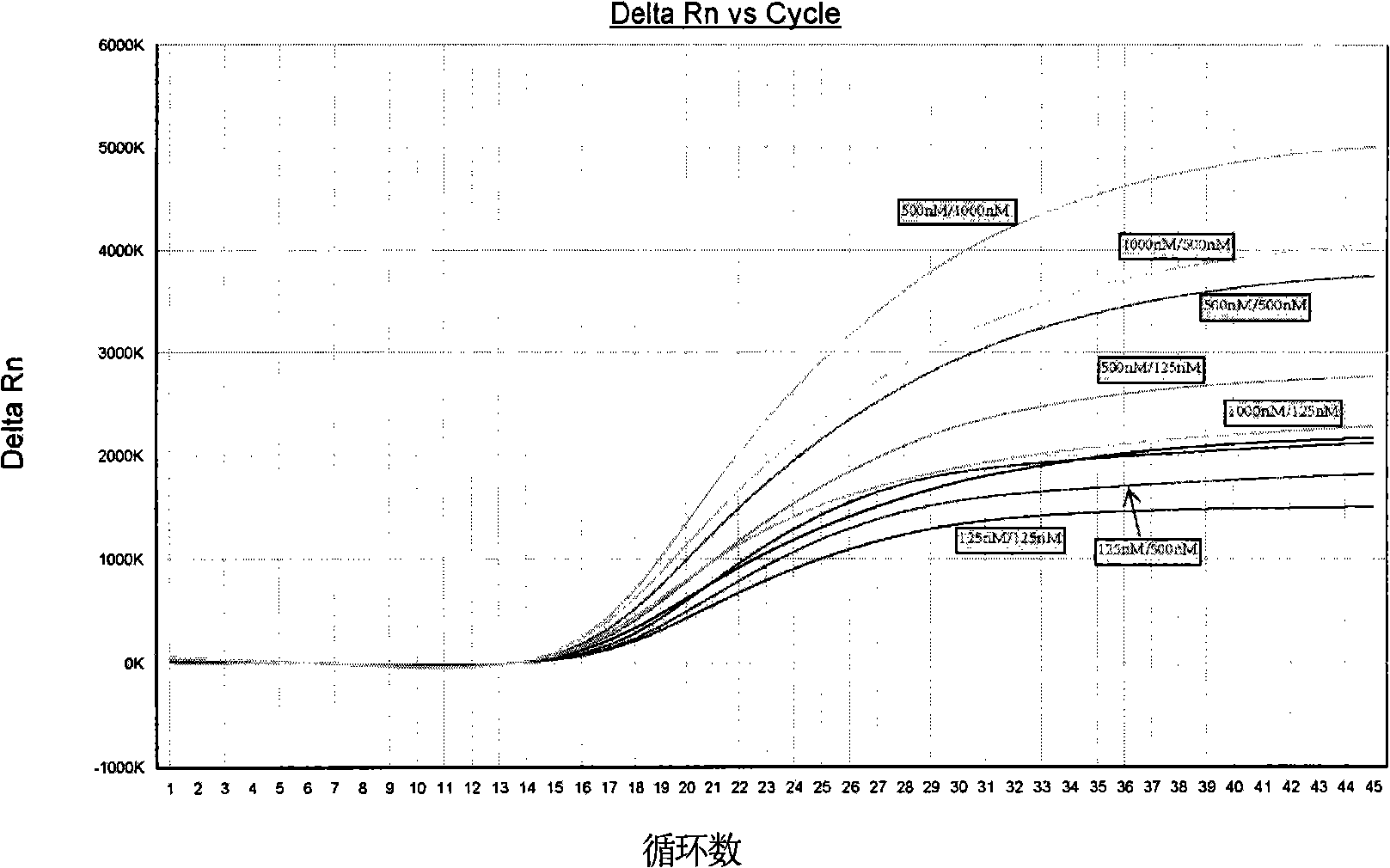
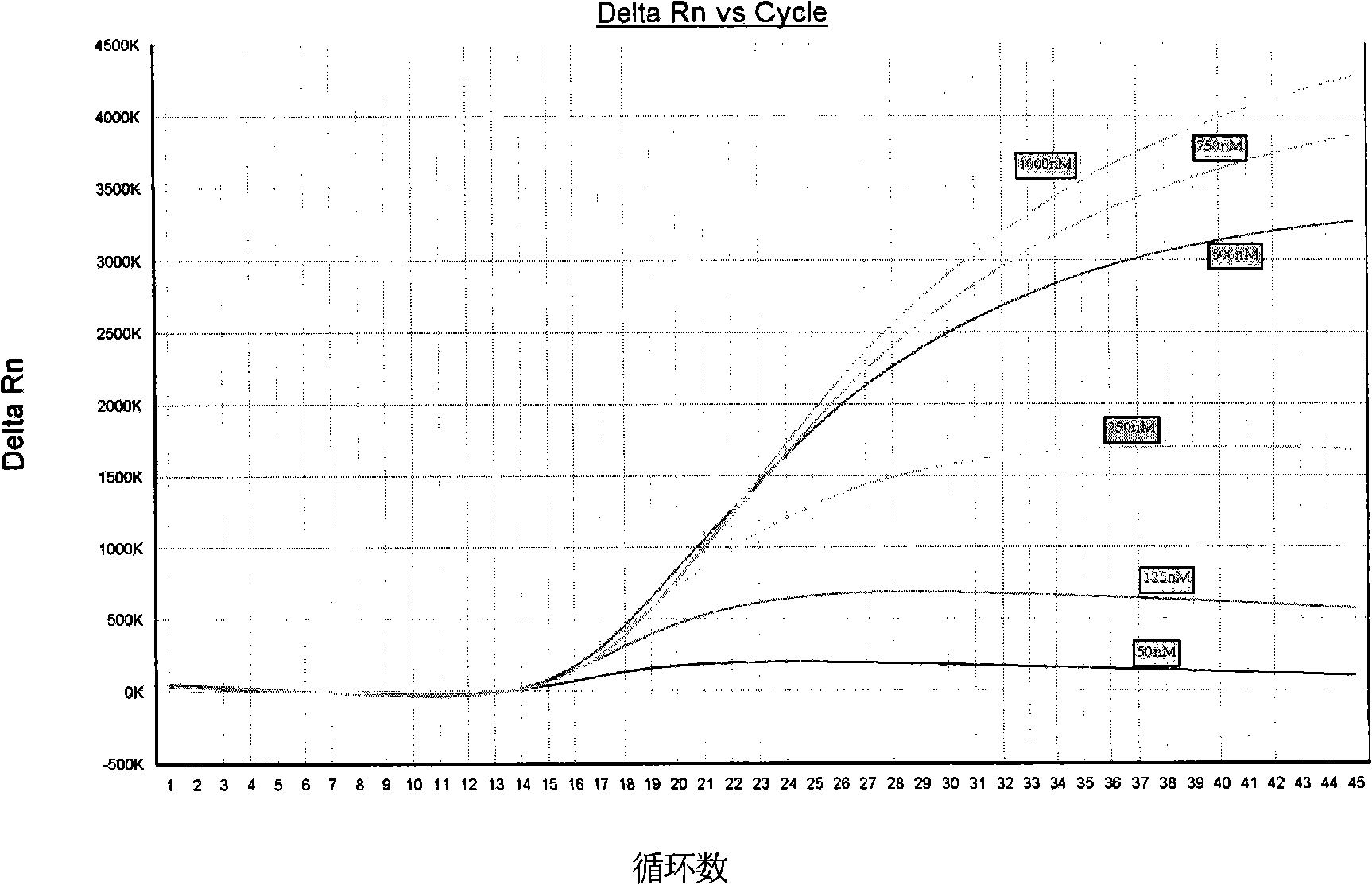
[0038] Embodiment 3, determination of amplification program
[0039] The kit used for TaqMan RT-PCR reaction is Ag-Path-ID TM One step RT-PCR Kit (product of Ambion, USA), amplification and detection were performed on Applied Biosystems 7500 Fast Real-Time PCR Systems, a fully automatic fluorescent quantitative instrument, or on instruments such as Stratagene Mx3005P real-time fluorescent quantitative PCR instrument.
[0040]According to the recommendations of the kit and the specific characteristics of the primers and probes, the reaction program on the instrument tested 3 temperatures and 3 times in the selection of annealing extension conditions: the reverse transcription reaction conditions were 50°C for 10 minutes, and 95°C hot start for 10 minutes. Minutes later, enter 45 two-step PCR cycles: 95°C for 10 seconds → (58 / 60 / 62)°C (30 / 40 / 60) seconds; ×RT-PCR buffer 10 μl, 20 μM forward primer CHIK-FP 1 μl, 20 μM reverse primer CHIK-RP 1 μl, 5 μM probe CHIK-Probe 0.5 μl, 25...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com