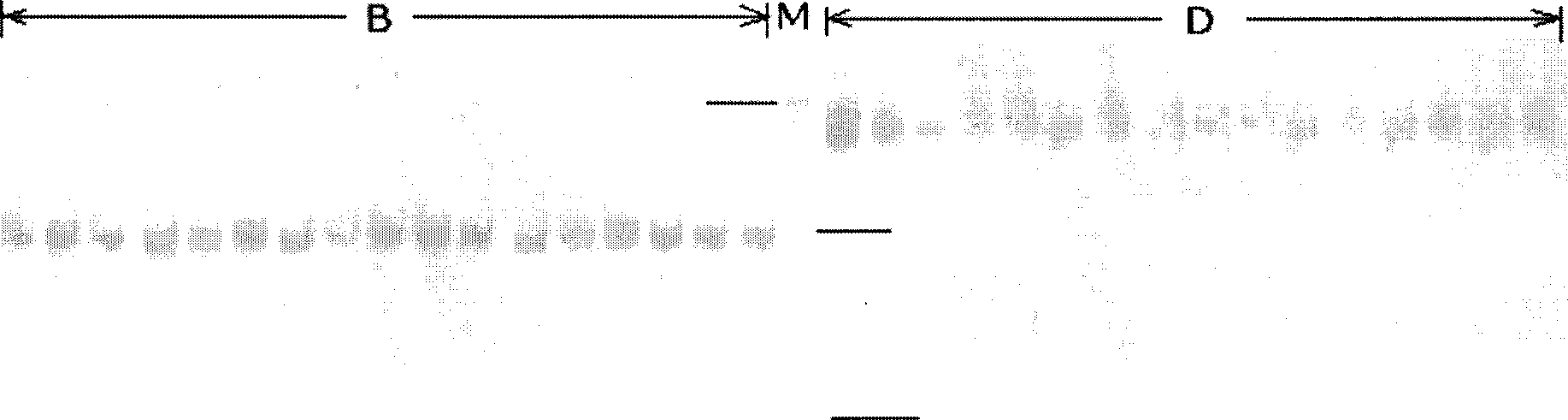Molecular identification method for siniperca kneri garman and siniperca scherzeri and kit
A technology for molecular identification and big-eyed mandarin fish, applied in biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of improving efficiency and accuracy, easy and fast operation, and wide applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] 1. Extraction of DNA:
[0027] Genomic DNA was extracted by the phenol-chloroform method. 0.8% agarose gel electrophoresis and nucleic acid protein analyzer (Eppendorf products) to detect DNA concentration and purity.
[0028] 2. Amplify DNA fragments:
[0029] That is, the polymerase chain reaction, the DNA sequence of a pair of primers for the polymerase chain reaction is:
[0030] 1) Forward primer: 5'GGCTCGTAGAAGCAGTAGGT 3',
[0031] Reverse primer: 5'GGGATAATGTAGTAGGTGGATT 3';
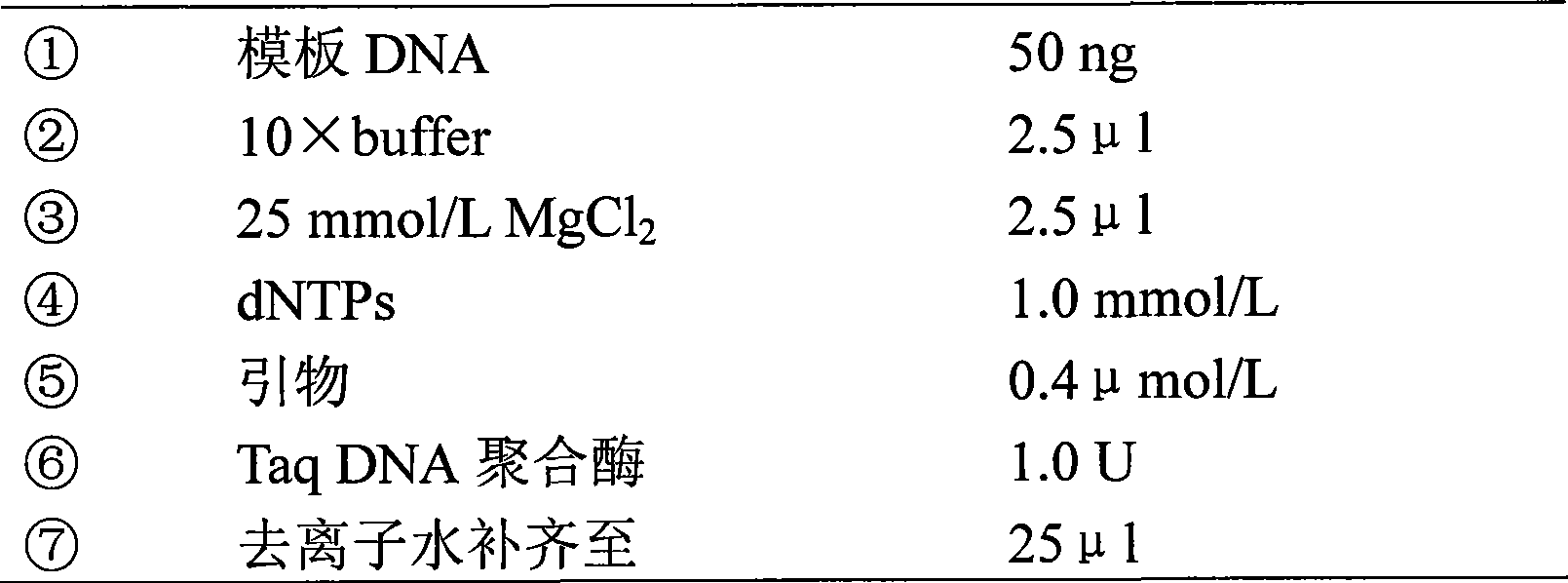
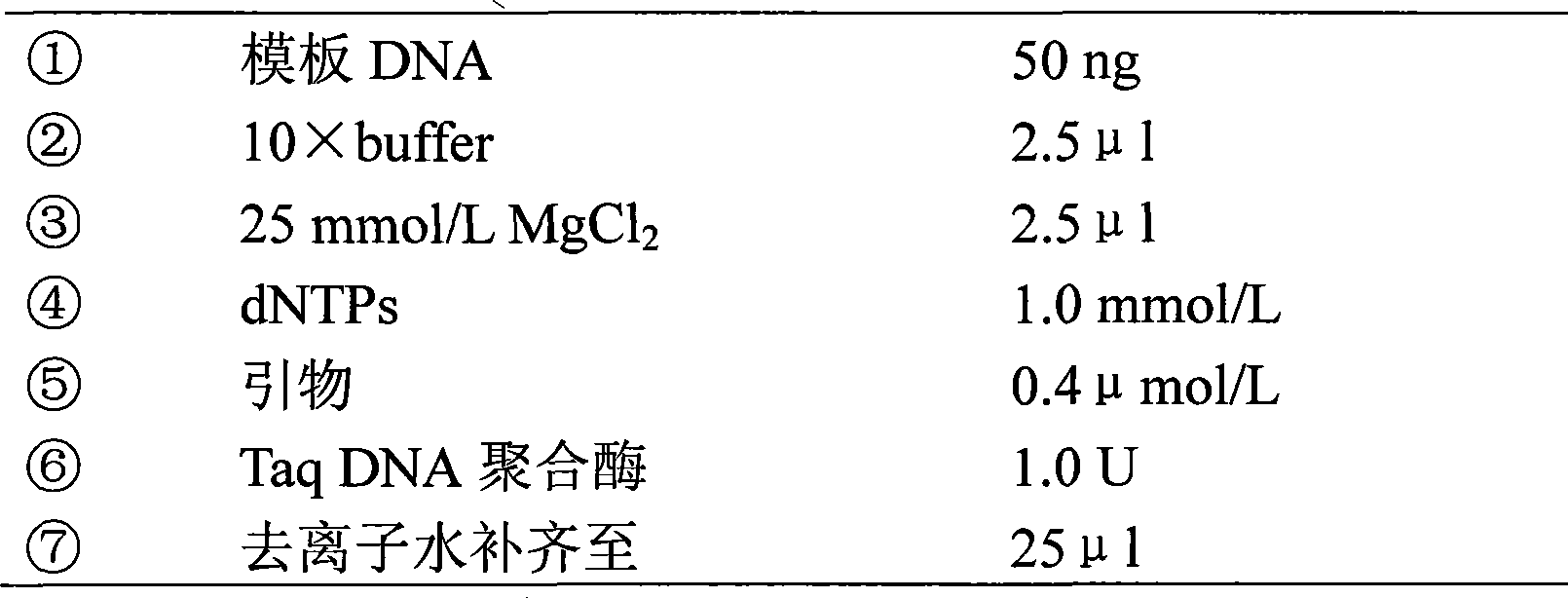
[0032] 2) The PCR reaction system is as follows:
[0033]
[0034] After mixing, centrifuge at high speed for a few seconds.
[0035] 3) PCR reaction conditions: The PCR reaction was carried out on a PCR machine, and the reaction program was: 94°C pre-denaturation for 5 minutes, and then 35 cycles according to the following conditions: 94°C denaturation for 30 seconds, 55°C annealing for 42 seconds, 72°C extension for 1 minute, cycle After the end of the reaction, extend at 72°C fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com