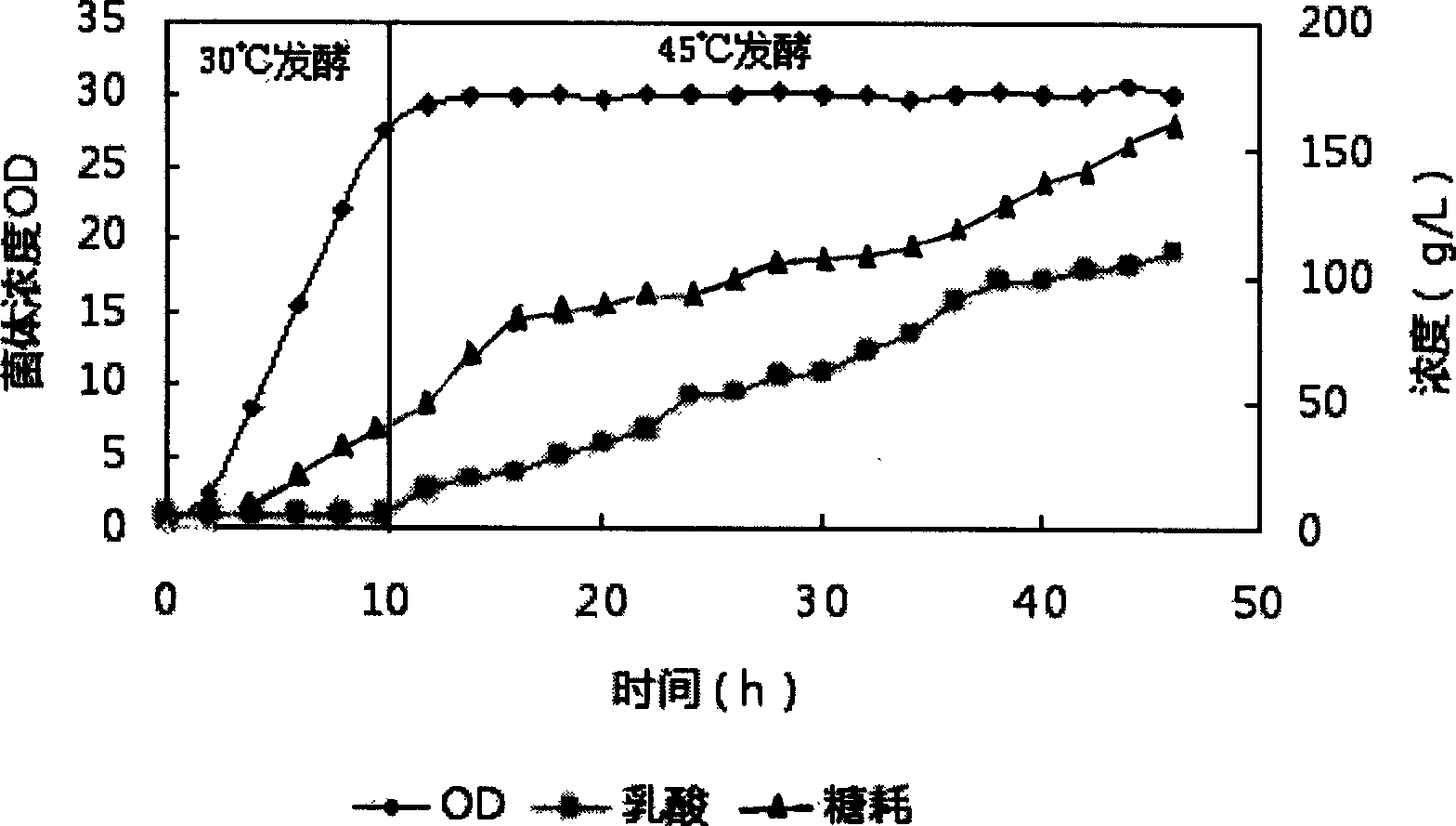
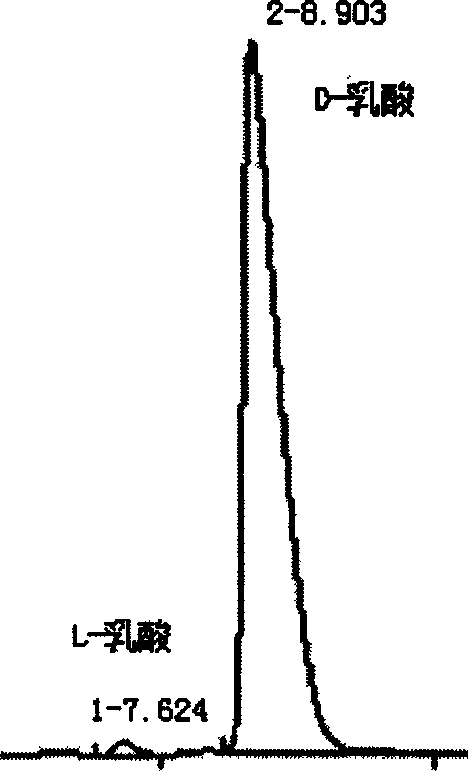
Method for preparing high optical purity D-lactic acid by fermentation
A technology of optical purity and lactic acid, which is applied in the field of microbial fermentation engineering, can solve the problem that acetic acid cannot be used as industrial lactic acid, and achieve the effect of ensuring the quality of polymerization and efficient synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 - Deletion of the gene encoding pyruvate dehydrogenase
[0034] Using synthetic oligonucleotide primers P1 and P2 as primers, P1 and P2 are:
[0035] P1: 5′-ATA GGATCC TATCGAAATCAAAGTACCGGACATCGGGG-3′
[0036] P2: 5′-CATCA GGATCC AGACGGCGAATGTCAGA-3′
[0037] Using Escherichia coli 947 chromosomal DNA as a template, the pyruvate dehydrogenase gene was amplified from the genome by PCR (polymerase chain reaction, Maties, 1989), and the PCR product was cloned into the BamHI site of pUC18 to obtain the recombinant plasmid pUC -PDH. Use EcoRV to remove the 963bp fragment and combine it with dif Ec -Gm R Ligation, to obtain the gene deletion sequence for pyruvate dehydrogenase, pdh′-dif-Gm R -dif-pdh', ie: pdh'::Gm R dif . This gene deletion sequence was transformed into E. coli. Transformants were selected for on selective media. Then subculture on non-selective medium, and select the transformant whose selectable marker disappears. The chromosomal D...
Embodiment 2
[0038] Example 2—Deletion of the gene encoding pyruvate formate lyase
[0039] Using synthetic oligonucleotide primers P3 and P4 as primers, P3 and P4 are:
[0040] P3: 5′-ATA GGATCC TGATTACCGCTGGCAACAACGA-3′
[0041] P4: 5′-CATCA GGATCC ATTGGCAACCAGGCAAGCGA-3′
[0042] Using Escherichia coli 947 chromosomal DNA as a template, the pyruvate formate lyase gene was amplified from the genome by PCR, and the PCR product was cloned into the BamHI site of pUC18 to obtain the recombinant plasmid pUC-PFL. Use EcoRV to remove the 1773bp fragment and combine it with dif Ec -Gm R Ligation, to obtain the gene deletion sequence for pyruvate formate lyase, pfl'-dif-Gm R -dif-pfl', ie: pfl'::Gm R dif . This gene deletion sequence was transformed into E. coli 947 (Δpdh). Transformants were selected for on selective media. Then subculture on non-selective medium, and select the transformant whose selectable marker disappears. The chromosomal DNA of the transformant was extracted, ...
Embodiment 3
[0043] Example 3—Deletion of the gene encoding FAD-dependent D-lactate dehydrogenase
[0044] Using synthetic oligonucleotide primers P5 and P6 as primers, P5 and P6 are:
[0045] P5: 5′-ATA GGATCC GCGATGTCTTCCATGACAACAACTG-3′
[0046] P6: 5′-CATCA GGATCC GGATTCATGCTGTTGGTCGGATC-3′
[0047] Using Escherichia coli 947 chromosomal DNA as a template, the pyruvate formate lyase gene was amplified from the genome by PCR, and the PCR product was cloned into the BamHI site of pUC18 to obtain the recombinant plasmid pUC-DLD. The 700bp fragment was digested with StuI and EcoRV and combined with dif Ec -Gm R Ligation, to obtain the gene deletion sequence of FAD-dependent D-lactate dehydrogenase, dld′-dif-Gm R -dif-dld', ie: dld'::Gm R dif . This gene deletion sequence was transformed into E. coli 947 (Δpdh, Δpfl). Transformants were selected for on selective media. Then subculture on non-selective medium, and select the transformant whose selectable marker disappears. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com