Real-time fluorescent quantitative polymerase chain reaction (PCR) detecting kit for coxsackievirus A16
A Coxsackie virus and kit technology, applied in the field of real-time fluorescent PCR detection kits, can solve problems affecting virus diagnosis and epidemic monitoring, and achieve the effect of good sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1: Coxsackievirus A16 fluorescent PCR detection kit and its use
[0036] 1. Prepare a kit including the following components: 1 tube of RNA extraction solution (50ml / tube), 24 tubes of PCR amplification reaction solution (20μl / tube), 1 tube of negative quality control (100μl / tube), 1 tube of positive quality control 1 tube of sample (100 μl / tube), 4 tubes of quantitative reference product (50 μl / tube).
[0037] 2. Specimen collection, transportation and storage
[0038] (1) Specimen collection: The object of specimen collection is clinically diagnosed cases of HFMD at the time of the outbreak and children under 5 years old in communities (villages) near the community (village) without cases or nursery institutions where the cases occurred (as healthy control population).
[0039] Specimens for virus isolation include feces, throat swabs, herpes fluid, and cerebrospinal fluid if the patient has neurological symptoms. Clinical specimens should avoid repeated...
Embodiment 2
[0067] Example 2: Preparation and use of quantitative reference and quality control in CA16 virus nucleic acid quantitative detection kit
[0068] The quantitative reference product in the CA16 virus nucleic acid quantitative detection kit is a plasmid containing the specific sequence of the CA16 virus nucleic acid, which is used to prepare a standard curve, accurately quantify the sample to be tested, and can be directly used for RT-PCR detection; quality control products include positive quality control The product and the negative quality control product are used for quality control in clinical trials, and the operation method is the same as that of the sample to be tested.
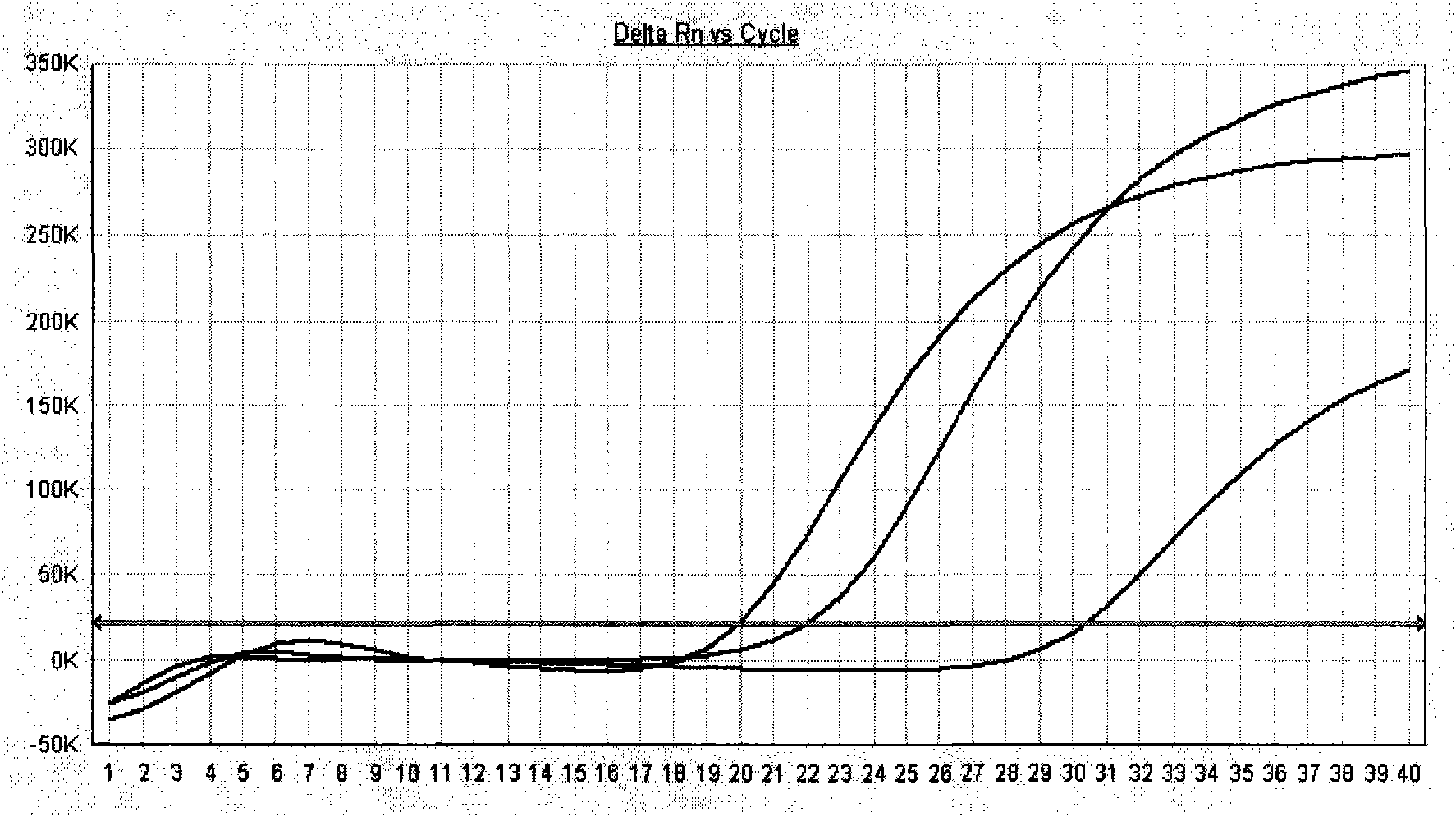
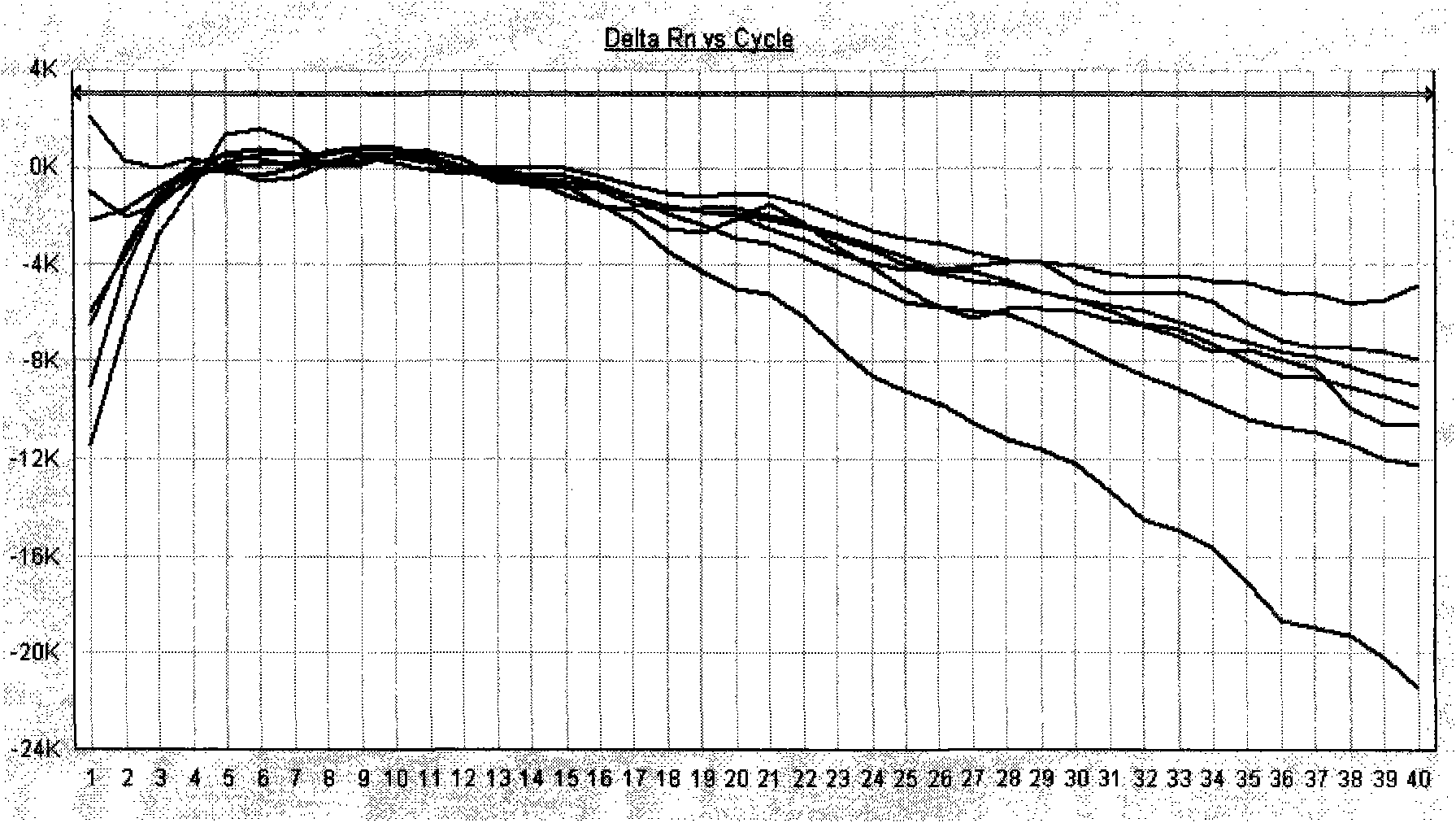
[0069] After one-step or two-step amplification, save the detection data file. Adjust the analysis parameters according to the analyzed image to optimize the standard curve graph under the standard curve (Std curve) window (that is, the correlation (correlation) value image 3 , and the information abou...
Embodiment 3
[0071] Example 3: Quantitative detection of clinical samples using the CA16 virus fluorescent PCR detection kit
[0072] The samples were from the Guangdong Provincial Center for Disease Control and Prevention, including feces, throat swabs, serum, cerebrospinal fluid and other sample types; the RNA extraction, PCR reaction and result analysis of the samples were carried out with reference to Example 1.
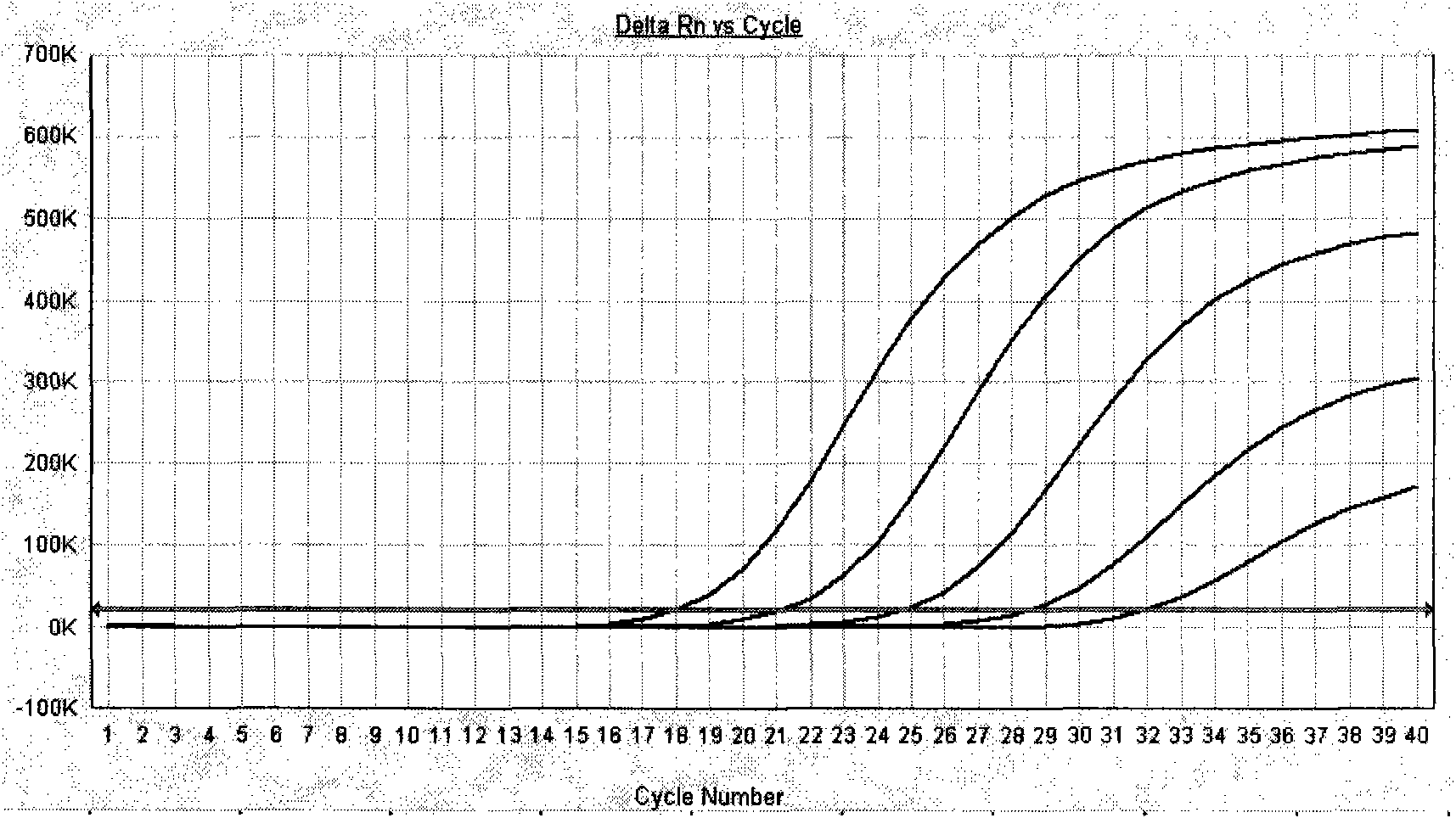
[0073] After the PCR reaction, adjust the analysis parameters according to the amplification curve to make the standard curve under the standard curve (Std curve) window reach the best (ie, the absolute value of the correlation value > 0.97), and then analyze the clinical samples. The test results of clinical samples are attached Figure 7 Shown: The Ct values of the amplification curves of the 5 clinically positive specimens are 24.35, 26.69, 27.67, 29.91, and 31.07, respectively. Combined with the obvious exponential growth period of the amplification curves, they can all...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com