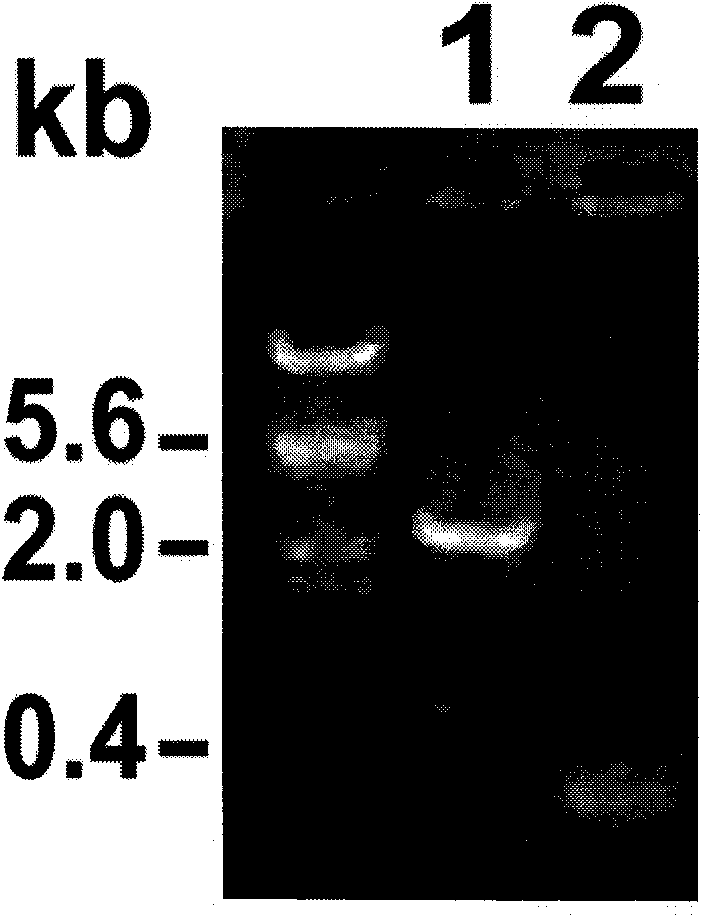Gene deletion system for security control of transgene monocotyledon
A monocotyledon, gene deletion technology, applied in the field of high-efficiency gene deletion system, can solve the problems of exogenous gene escape, poor operability, and low efficiency of exogenous gene
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] [Example 1] Construction of plant expression vector
[0047] 1. Cloning of maize pollen-specific promoter ZM13
[0048] According to the report of Hamilton et al. (Hamilton D.A.et al, Sex.Plant Reprod.1989, 2, 208-212), two pairs of primers were synthesized respectively. The upstream primers were: 5'>GCGGTACCCCACTTCGCGGATTC CACGTCTGCAGTATGGTACCAACGGCGGC CC<3' (PstI), the pollen-specific promoter ZM13 was amplified by PCR from the maize (Zea mays) genome.
[0049] React component:
[0050] Genomic DNA (25ng / μL) 2 microliters
[0051] dNTP (10mM each) 2 microliters
[0052] Magnesium chloride (5mM) + 10 times buffer 2 microliters
[0053] Upstream primer, downstream primer (each 100μM) 2 microliters
[0054] Platimum Pfx DNA Polymerase 2U
[0055] DNA polymerase 10x buffer 5 μl
[0056] Total reaction volume 50 μl
[0057] Reaction parameters: 95°C, 5 minutes. 94°C for 1 minute, 57°C for 1 minute, 72°C for 1 minute, 35 cycles. Extend at 72°C for 10 min.
[0...
Embodiment 2
[0079] [Example 2] Obtaining of transgenic corn plants
[0080] genetic transformation
[0081] Genetic transformation of maize see Wang et al. ( http: / / www.agron.iastate.edu / ptf / service / biolisticmaize.aspx) method. Take the immature embryos of maize Hi II line (from A188×B73) as explants, soak them in 70% ethanol for 2 min after removing the bract leaves, and then soak them in 0.1% HgCl 2 Disinfect in the solution for 20 minutes, rinse with sterile water 3 times, blot dry the water stains, and inoculate on the induction medium. Add 2mg / L 2,4-D to MS medium to induce embryogenic callus.
[0082] Embryogenic calli at different growth stages were used for transformation. The model of the gene gun is BiolisticPDS-1000 / He Particle Delivery System (Bio-Rad). The bombardment parameters are: the distance between the cleavable disk and the carrier is 2.5cm, the distance between the carrier and the barrier net is 0.8cm, the distance between the barrier net and the target cell is...
Embodiment 3
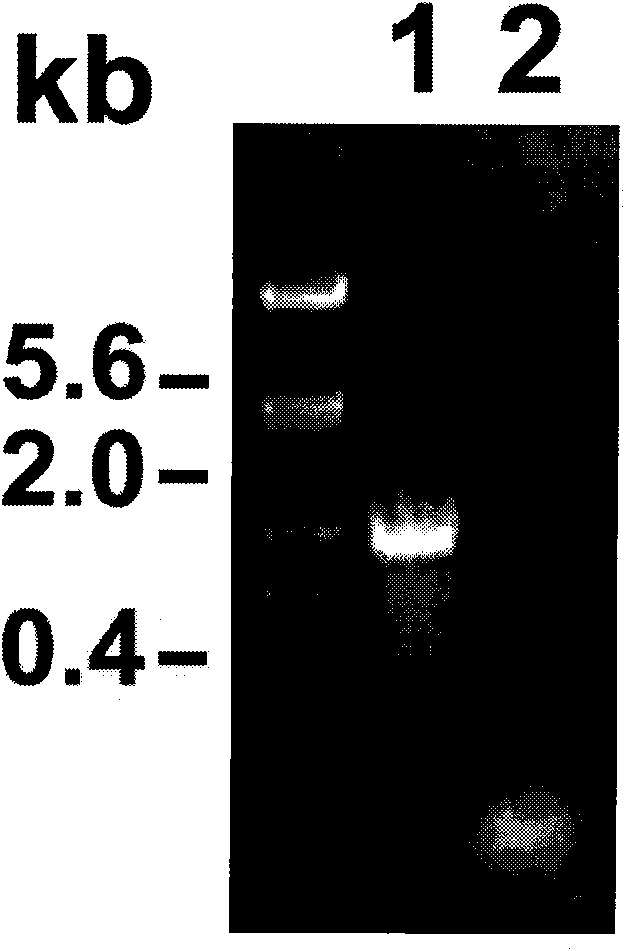
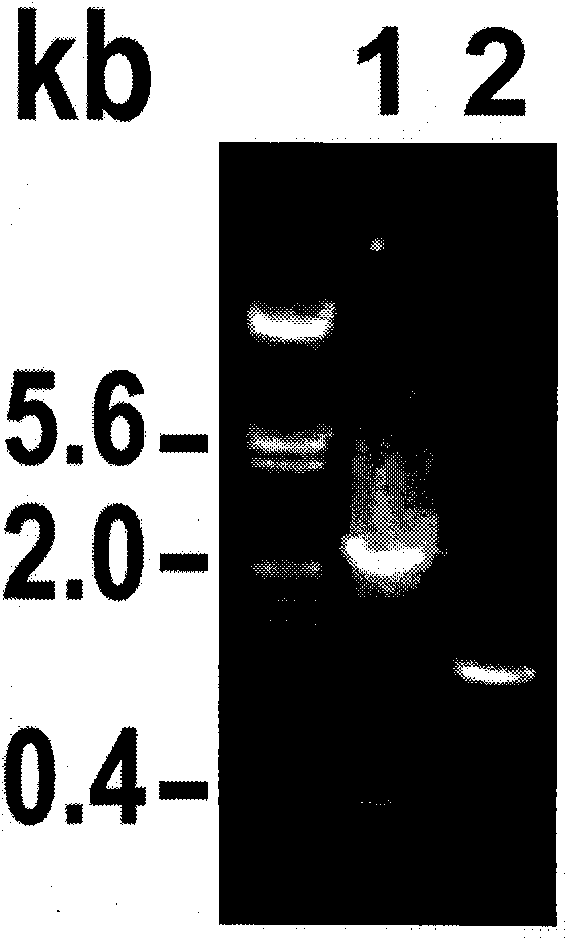
[0094] [Example 3] Expression Analysis of Exogenous Genes in Transgenic Plants
[0095] 1. Expression analysis of GUS gene in transgenic pLF-ZM13-FLP-Ubi-GN maize
[0096] The pollen-specific promoter ZM13 was selected to control the expression of the site-specific recombinase FLP gene. In theory, in transgenic pLF-ZM13-FLP-Ubi-GN plants, the recombinase FLP gene controlled by the ZM13 promoter should only be expressed in mature pollen Expressed in, delete the foreign gene. Therefore, abundant expression of the GUS reporter gene (controlled by the constitutive promoter Ubi) should be detectable in all tissues before pollen maturation in transgenic plants. In order to detect whether the promoter ZM13 has tissue-specific expression characteristics, T 0 The roots, stems and leaves of the generation plants were stained with GUS tissue, Figure 8 Shown in is the GUS staining results of different tissues of pLF-ZM13-FLP-Ubi-GN transgenic plants. It shows that the exogenous gene ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com