Microorganism capable of producing l-amino acid, and method for production of l-amino acid
A technology of microorganisms and amino acids, applied in the fields of peptides, DNA/RNA fragments, hydrolases, etc., can solve problems such as no research yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
reference example 2
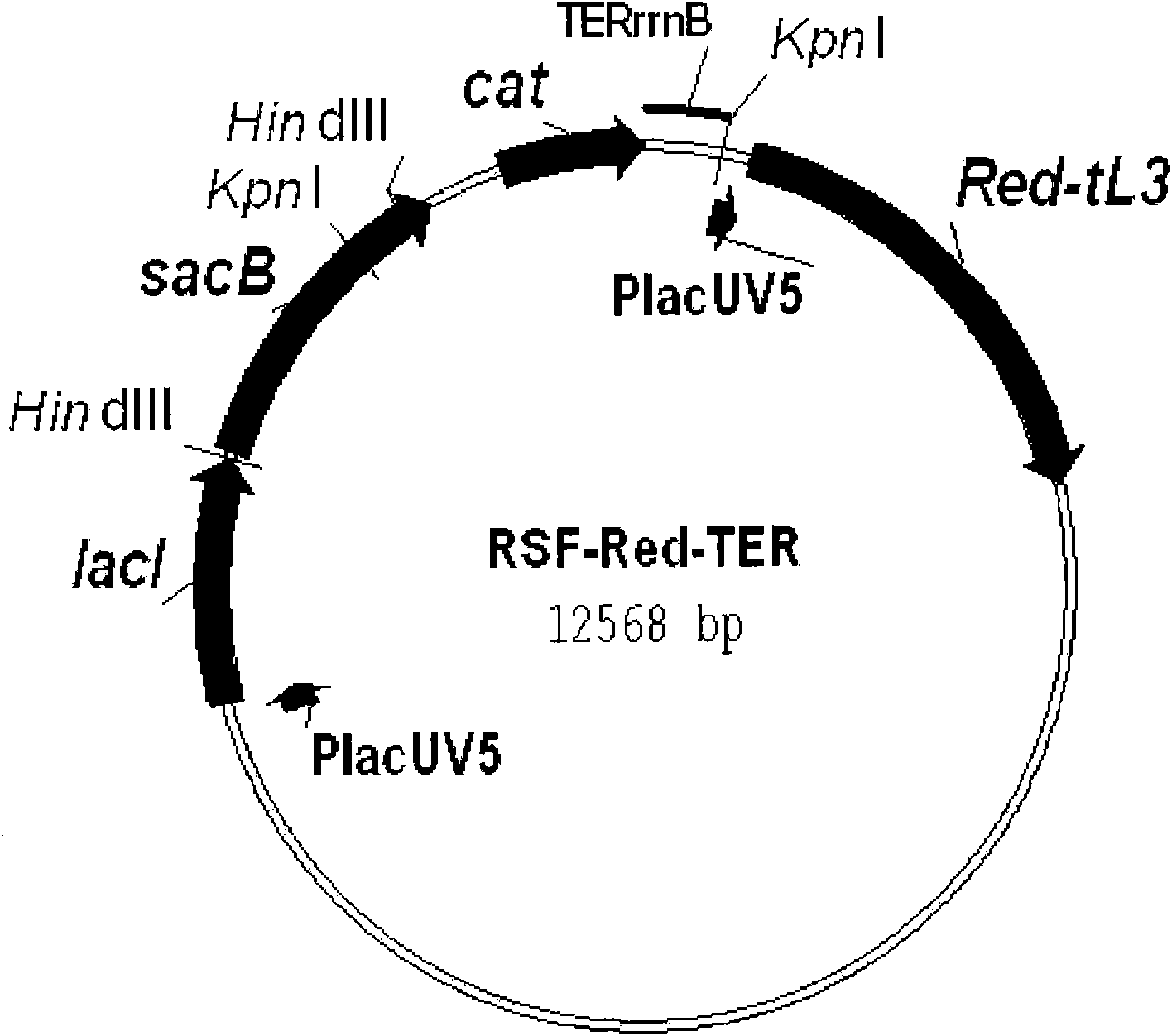
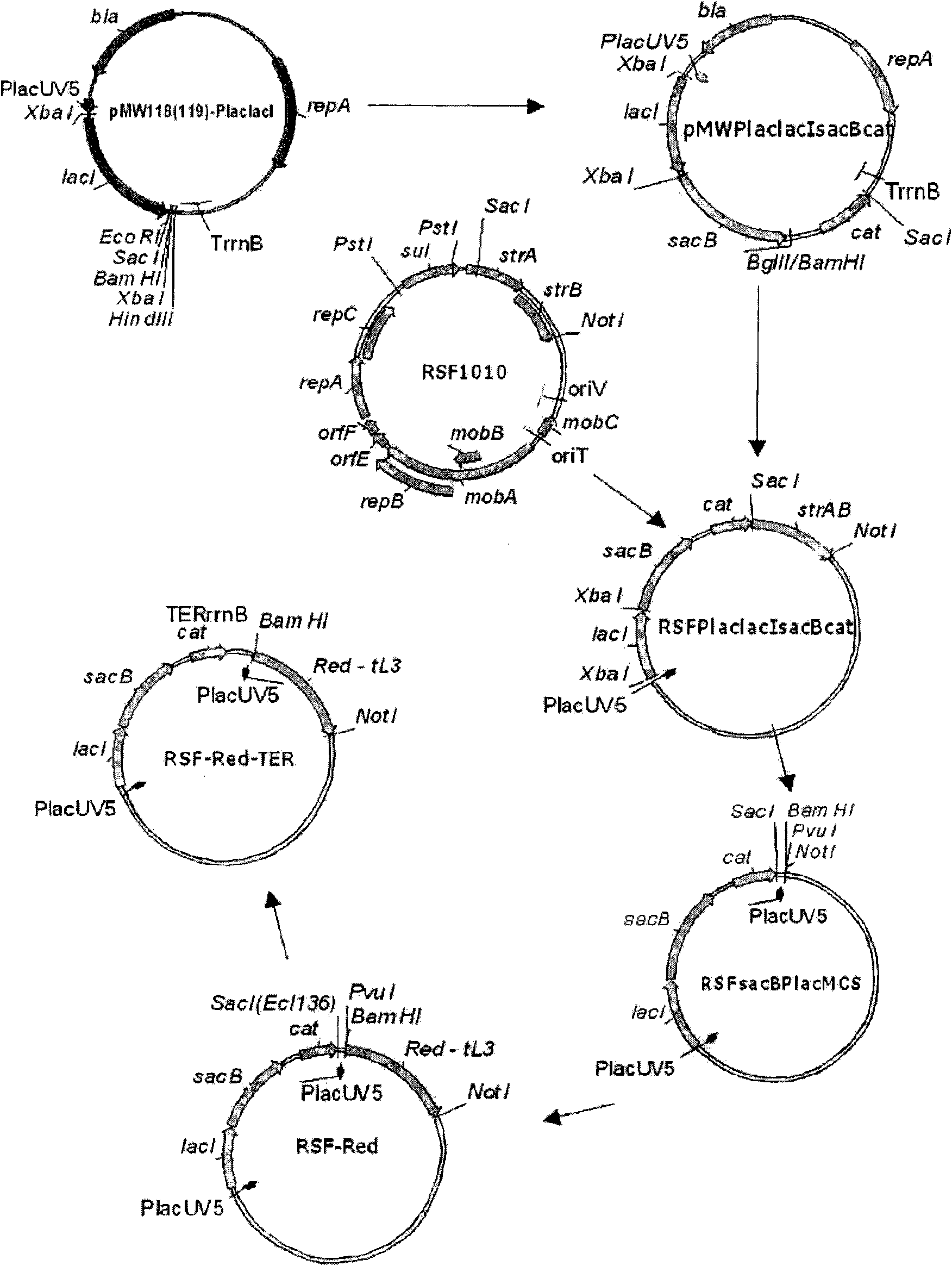
[0226] [Reference Example 2] Construction of the helper plasmid RSF-Red-TER
[0227] The construction scheme of the helper plasmid RSF-Red-TER is as follows: figure 2 shown.
[0228] As an initial step of construction, the RSFsacBPlacMCS vector was designed. To this end, DNA fragments comprising the cat gene of the pACYC184 plasmid and the structural part of the sacB gene of Bacillus subtilis were amplified by PCR using oligonucleotides of SEQ ID NO: 17, 18, 19, 20, respectively. The 5' ends of these oligonucleotides contained BglII, SacI, XbaI, and BamHI restriction enzyme sites, respectively, necessary and convenient for further cloning. The resulting 1.5kb sacB fragment was cloned into the previously obtained pMW119-P lac XbaI-BamHI site of the lacI vector. The vector is in accordance with pMW118-P lac The related description of lacI vector (Skorokhodova, A. Yu et al., Biotekhnologiya (Russian), 5, 3-21 (2004)) was constructed by the same method. Only this vector con...
Embodiment 1
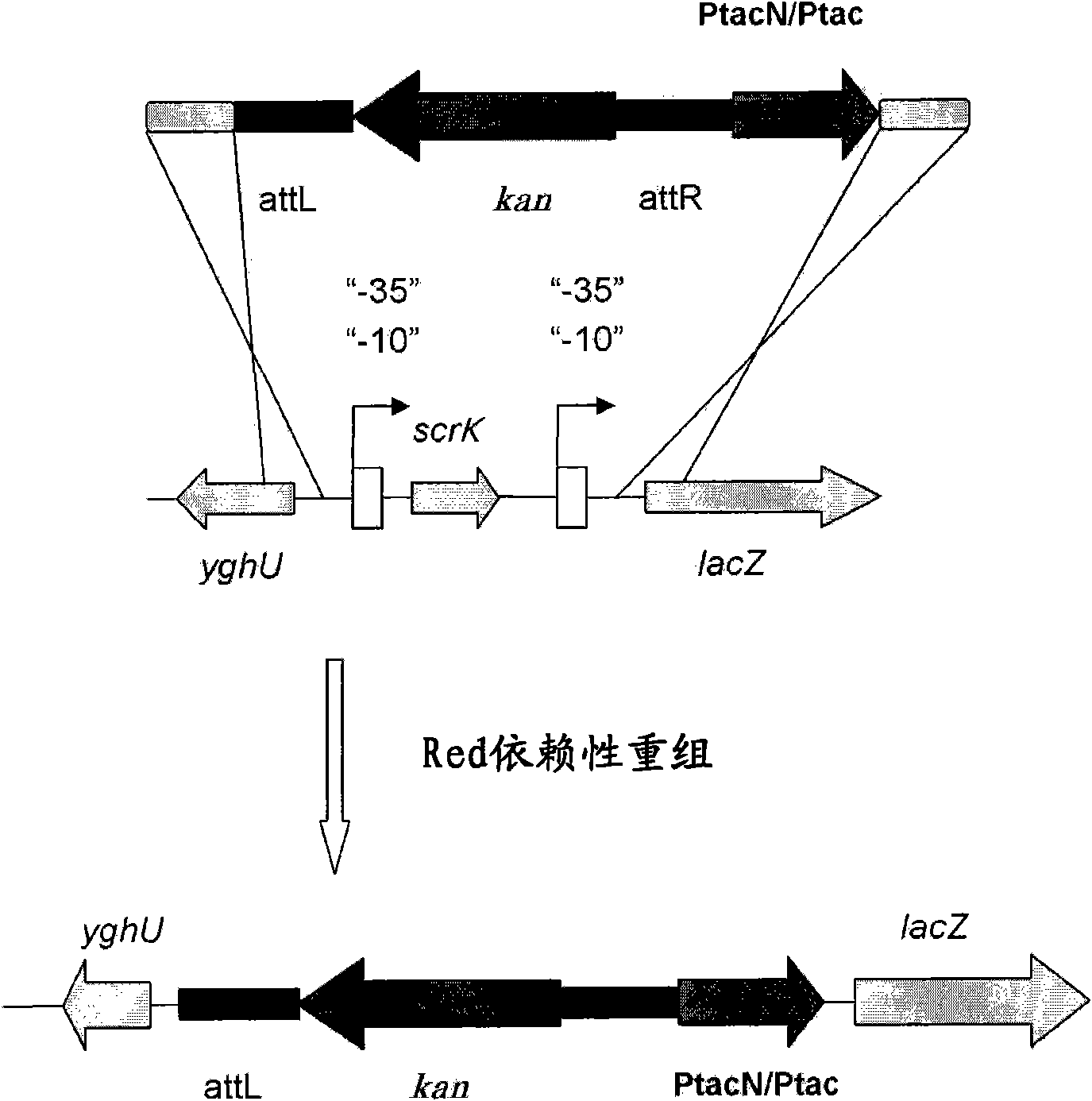
[0242] [Example 1] Acquisition of promoter substitution strain of kdp operon
[0243] (1) Construction of glutamic acid production plasmid RSFPPG
[0244] A plasmid RSFPPG in which L-glutamic acid biosynthetic system gene, prpC gene (International Publication No. 2006 / 051660 pamphlet), ppc gene, and gdh gene (European Application Publication No. 0999282 pamphlet) was amplified was constructed.
[0245] Primer 1 (SEQ ID NO: 40) and primer 2 (SEQ ID NO: 41) were designed to amplify parts other than the ORF of the gltA gene of RSFCPG (European Application Publication No. 1233068 specification). Using these primers, PCR was performed using RSFCPG as a template to obtain a fragment of about 14.9 kb. On the other hand, for prpC, PCR was performed using primer 3 (SEQ ID NO: 42) and primer 4 (SEQ ID NO: 43) using the chromosomal DNA of Escherichia coli W3110 strain as a template, and a fragment of about 1.2 kb was obtained. The two PCR products were treated with BglII and KpnI respe...
Embodiment 2
[0325] [Example 2] Amplification of the kdp operon in L-threonine-accumulating Escherichia coli
[0326] (1) Construction of a plasmid for amplification of the kdp operon
[0327] In order to introduce the kdp operon into bacteria belonging to the genus Escherichia, a plasmid for amplifying the kdp operon was constructed using the known plasmid pMW218 (Takara Shuzo Co., Ltd.).
[0328] First, pMW218 was digested with restriction enzymes HindIII and BamHI, and a phenol-chloroform solution was added and mixed to stop the reaction. After the reaction solution was centrifuged, the upper layer was recovered, and DNA was recovered by ethanol precipitation. On the other hand, using the chromosome extracted from Escherichia coli MG1655 as a template, PCR was carried out using DNA primers shown in SEQ ID NO: 55 and 56 (denaturation at 94° C. for 10 seconds, annealing at 60° C. for 30 seconds, extension reaction for 72 °C - 120 sec) to amplify the kdp operon. Pyrobest DNA polymerase ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com