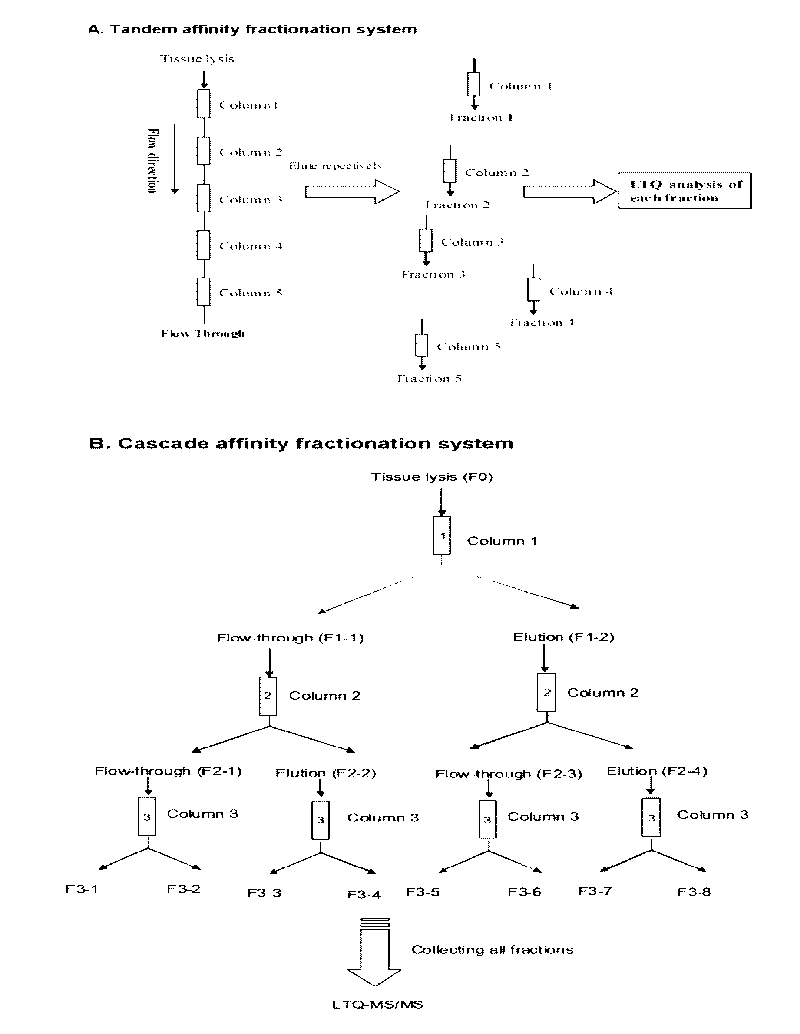
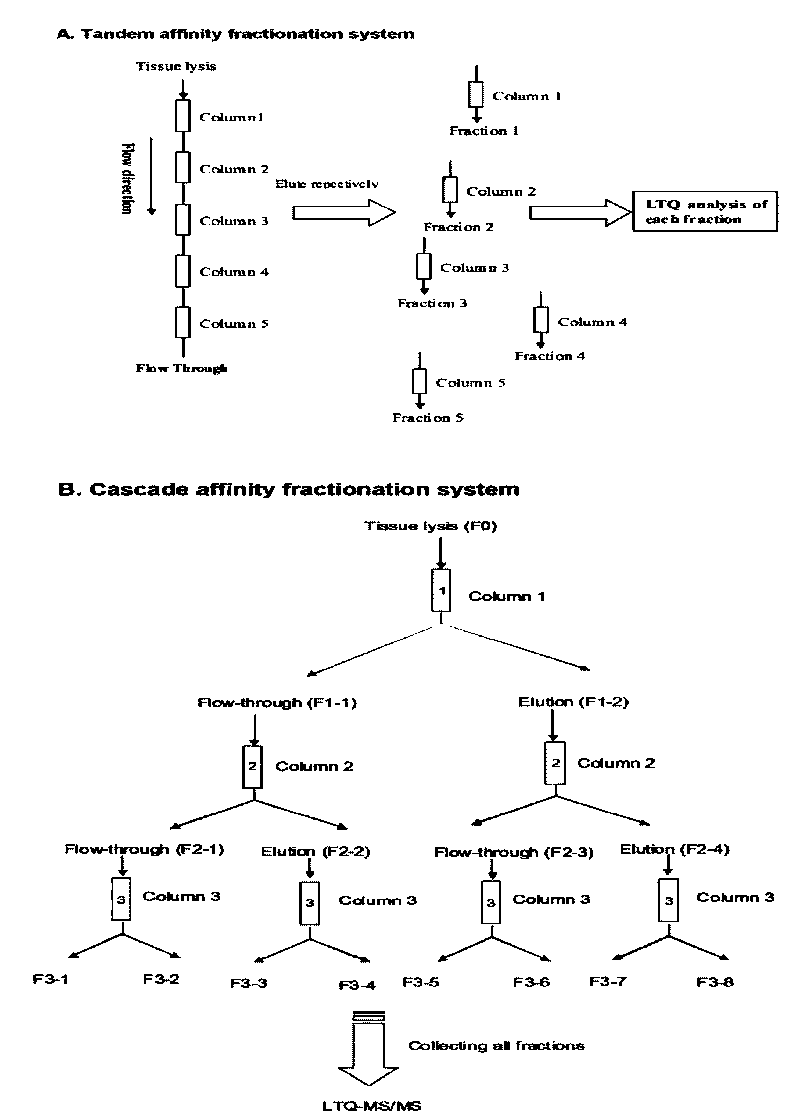
Method for utilizing affinity column to analyze holoprotein
A whole protein and affinity technology, applied in the field of proteomics, can solve the problems of heavy workload, long operation time, time-consuming and laborious, and achieve the effect of increasing the number of proteins, simplifying the complexity, and being easy to automate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Synthesis of affinity materials based on epichlorohydrin
[0030] Take Sepharose CL-4B (100 grams) and wash it with 10 times the volume of deionized water, drain it into a wet cake; then suspend it in 50ml activation buffer (1M NaOH, 2.5g sodium borohydride, 10ml epichlorohydrin), React at a constant temperature of 60°C for 2 hours under stirring, stop the reaction when the pH is close to 7.0, then pour it into a glass frosted funnel, wash with 10 times the volume of distilled water under suction filtration, and distribute the Sepharose 4B activated by epichlorohydrin in a can In a sealed glass bottle (20g / bottle).
[0031] Take 0.5g of various amino compounds (see Table 1) and dissolve them in 25mL of 0.1M sodium hydroxide / L dioxane, respectively add them to the bottles containing activated Sepharose 4B, label them well, and put them under stirring at 60 ℃ reaction 24h. Add 1mL of mercaptoethanol, continue to react for 2 hours, fully wash with 10 times the volume of ...
Embodiment 2
[0077] Synthesis of Biomimetic Ligands Using Trichlorotriazazine as Skeleton
[0078] Suspend 100 grams of epichlorohydrin-activated Sepharose CL 4B medium in 350 ml of deionized water, then add 150 ml of 35% (v / v) ammonia water, and keep the temperature at 30° C. for 12 hours under stirring (200 r / min). NH 2 - Suspend SepharoseCL-4 B in 350ml of 50% (v / v) ice-bathed acetone solution, then dissolve 8g of trichlorotriazoxide in 80ml of -20°C pre-cooled acetone, quickly add the medium suspension, wash with saturated NaHCO 3 Keep the pH between 6.5 and 8.0, and continue stirring at 0 to 4°C for 2 hours to stop the reaction.
[0079] Use 3 times of medium volume of acetone, acetone: deionized water (volume ratio 1: 1), deionized water, wash the reaction in sequence to obtain dichlorotriazine-amino-Sepharose CL 4B; weigh 20g of dichlorotriazoxide -Amino-Sepharose CL 4B, mixed with a 5-fold molar excess of amino compound (R1) dissolved in a certain amount of distilled water (20-50...
Embodiment 3
[0081] Protein adsorption performance evaluation and selection
[0082] Evaluate the adsorption protein performance of the affinity separation material library, and select an affinity column whose adsorption protein species account for 20 to 80% of the total protein.
[0083] First, take 1ml of the synthesized ligand medium and fill it into the chromatography column respectively, write the label, and fully wash the balance with 15ml of equilibrium buffer (pH7.0, 10mM phosphate buffer containing 0.1M NaCl); then take Load 1-4mg tissue protein, combine on the column for 30 minutes, wash off unbound protein with 10-20ml equilibration buffer (monitored by protein UV detector until the baseline is flat); finally use 3ml elution buffer (pH12, 10mM Glycine-NaOH buffer) to elute the bound protein on the column and immediately adjust the pH to neutral. The collected protein samples bound by each ligand were analyzed by SDS-PAGE electrophoresis, and the ligand media with large differen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| cover factor | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com