Kit for detecting coxsackie virus A16-type nucleic acid and detection method thereof
A Coxsackie virus and detection kit technology, applied in biochemical equipment and methods, microbial measurement/inspection, fluorescence/phosphorescence, etc., can solve problems such as high time consumption, inability to meet detection requirements during outbreaks, and cross-contamination , to achieve the effect of easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0057] (2) Preparation of in vitro transcription RNA quality control products
[0058] After digesting the above-mentioned extracted plasmid with SAC I enzyme from Takara Company to make it linearized, the T7 RNA polymerase from Takara Company was used to perform in vitro transcription on the above-mentioned enzyme digestion reaction solution, and the obtained in vitro transcription reaction solution was processed with DNase I from Takara Company. Digestion was carried out to remove the plasmid DNA in it, and then the RNA in the digestion solution was extracted with TransZol of Quanshijin Company, that is, the RNA was transcribed in vitro; after it was quantified with a UV spectrophotometer, it was diluted to a concentration of 10 5 copies / μL was used as a quality control for in vitro transcription RNA.
[0059] The sequence of the Coxsackie type A16-specific target fragment inserted in the recombinant plasmid is shown in SEQ ID NO.4 in the sequence listing:
[0060] TTGCAGAC...
Embodiment 1
[0066] Method for detecting Coxsackievirus A16 type nucleic acid on ABI 7300 fluorescent quantitative PCR instrument with kit of the present invention
[0067] (1) RNA extraction: using QIAamp Viral RNA Mini Kit to extract,
[0068] (2) PCR Mix preparation: according to the ratio of 22 μL: 1 μL, draw the PCR reaction liquid and PCR reaction enzyme respectively, mix thoroughly, centrifuge at 12,000 rpm for 10 seconds, and take out the PCR reaction tubes, one of which is a positive quality control product reaction tube, and the other is Negative quality control product reaction tubes, and the rest are test sample reaction tubes, add 23 μL of PCRMix prepared above to each reaction tube;
[0069] The PCR Mix preparation method is shown in the table below:
[0070]
[0071] (3) Adding samples: add 2 μL of extracted sample RNA, 2 μL of positive quality control substance and 2 μL of negative quality control substance to the three reaction tubes, mix well and centrifuge at 12,000 ...
Embodiment 2
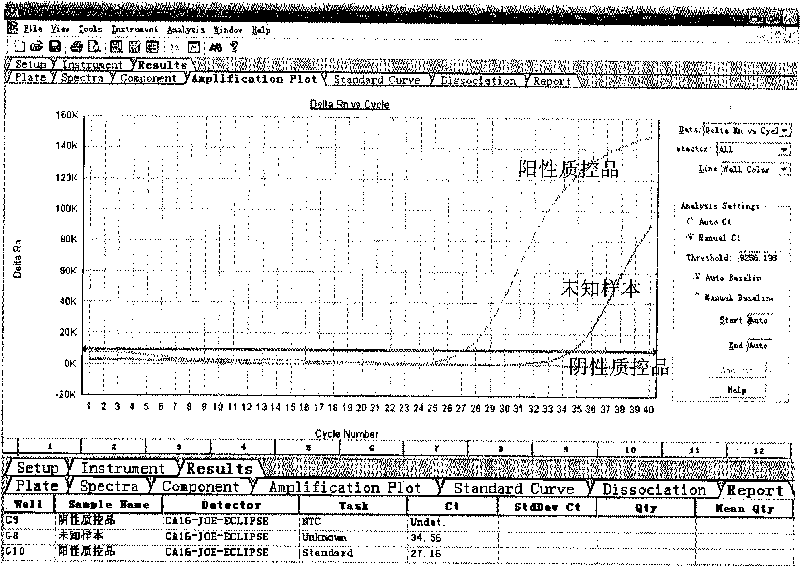
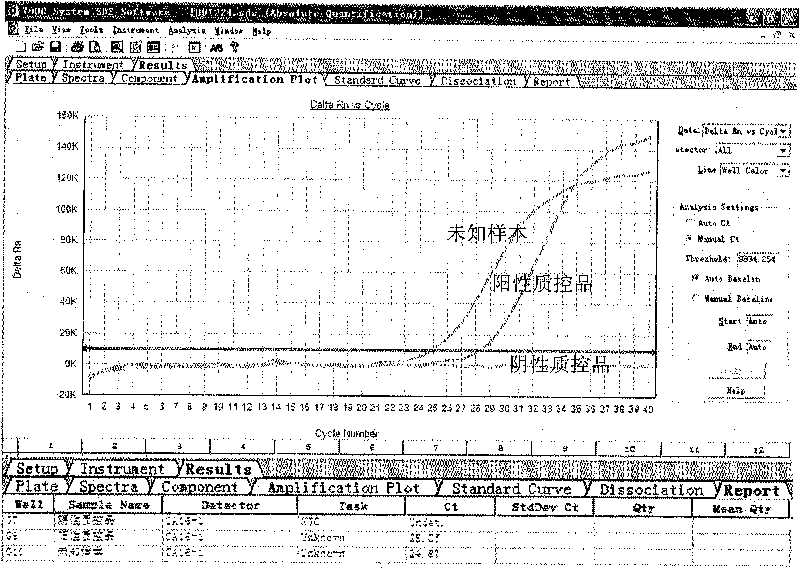
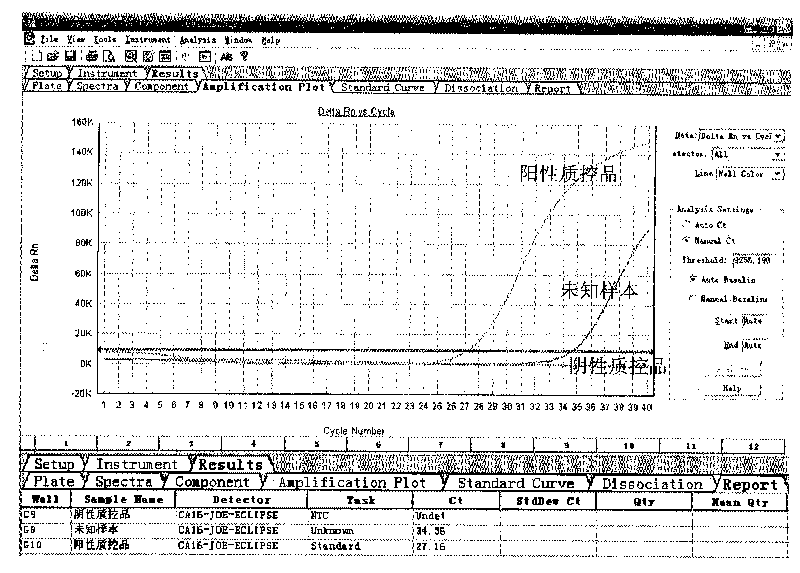
[0091] Use the test kit of the present invention to detect unknown samples according to the method of embodiment 1, and the unknown samples are derived from samples to be determined by patients in XX hospital. The test results of embodiment 2 of the present invention are as follows: figure 2 As shown, the test result was positive, and the Ct value was 24.87.
[0092] The embodiments described above are only one of the more preferred specific implementations of the present invention, and the usual changes and replacements performed by those skilled in the art within the scope of the technical solutions of the present invention should be included in the protection scope of the present invention.
[0093] CA16-1 Sequence List_ST25.txt
[0094] SEQUENCE LISTING
[0095] Beijing Aipuyi Biotechnology Co., Ltd.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com