Pathogen detection method
A detection method and technology for pathogenic bacteria, which are used in the determination/inspection of microorganisms, biochemical equipment and methods, and resistance to vector-borne diseases, etc. problem, to achieve the effect of rapid identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Preparation of multivalent phage M13KE-TC
[0025] Preparation of tetracysteine TC fragment:
[0026] Two TC DNA complementary sequences with Acc65 I and Eag I cohesive ends were designed and synthesized in Sansheng.
[0027] P1: 5'-GTACCTTTTCTATTCTCACTCTTTTCCTGAACTGTTGTCCCGGCTGCTGCATGGAGCCTTC-3' (denoted as SEQ ID NO.1);
[0028] P2: 5'-GGCCGAAGGCTCCATGCAGCAGCCGGGACAACAGTTCAGGAAAGAGTGAGAATAGAAAG-3' (denoted as SEQ ID NO. 2).
[0029] Take P1 and P2 respectively and mix them in Taq Buffer at a final concentration of 10M, and perform annealing reaction in a PCR instrument. The setting program is as follows: 95°C for 2min; continuously lower to 25°C for about 60min; store at 4°C.
[0030] The 7222bp multivalent phage M13KE DNA was recovered by double digestion with Eag I and Acc65 I. After the double digestion fragment was ligated with the annealed TC DNA fragment with sticky ends of Eag I and Acc65 I, it was transfected into E. coli TG 1. In the state, pla...
Embodiment 2
[0051] Example 2 Preparation of monovalent phage M13KO7-TC
[0052] 1. Construction and identification of recombinant phagemid pCANTAB 5E-TC
[0053]Two TC DNA complementary sequences with Not I and Sfi I cohesive ends were designed and synthesized in Sansheng.
[0054] P3: 5`-CGGCCATTCCTGAACTGTTGTCCCGGCTGCTGCATGGAGCCTGC-3` (denoted as SEQID NO.3);
[0055] P4: 5`-GGCCGCAGGCTCCATGCAGCAGCCGGGACAACAGTTCAGGAATGGCCGGCT-3`
[0056] (Denoted as SEQ ID NO.4). Take P3 and P4 respectively and mix them in Taq Buffer at a final concentration of 10 M, and perform annealing reaction in a PCR instrument. The setting program is as follows: 95°C for 2 minutes; continuously lower to 25°C for about 60 minutes; store at 4°C.
[0057] The 5.3kb phagemid DNA pCANTAB 5E was digested with Not I and Sfi I, recovered, and the double digested fragment was ligated with the annealed TC DNA fragment with Not I and Sfi I sticky ends to construct a recombinant phagemid. Cloning and sequencing were carri...
Embodiment 3 4
[0083] Example 3 Infection and double arsenic dye staining of specific pathogenic bacteria by tetracysteine recombinant bacteriophage
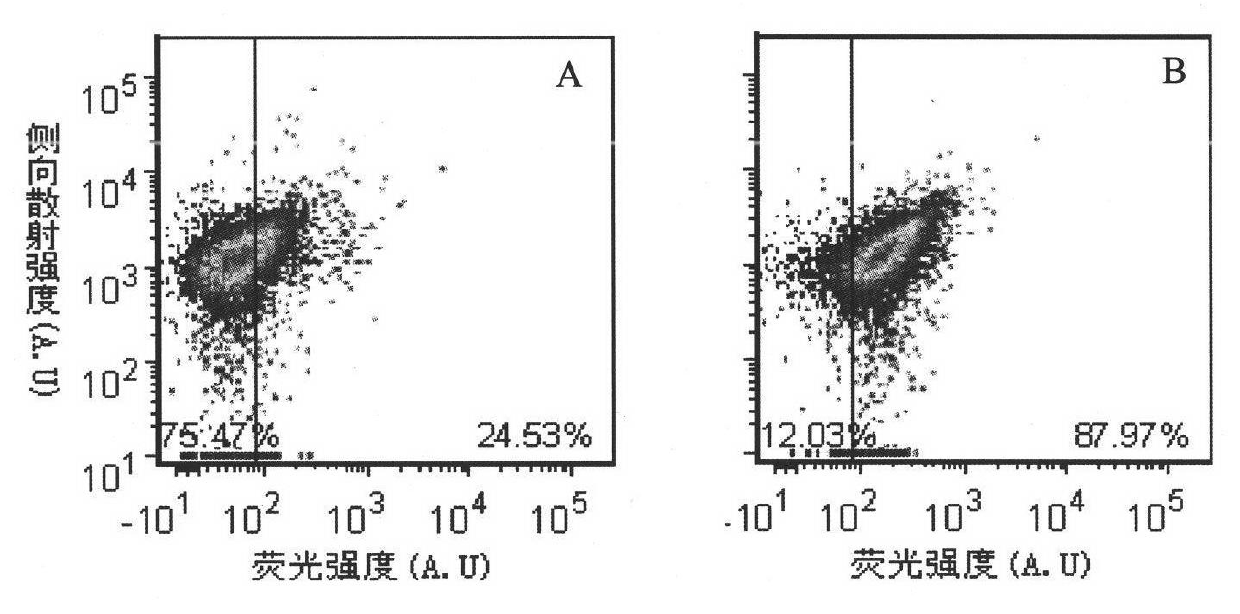
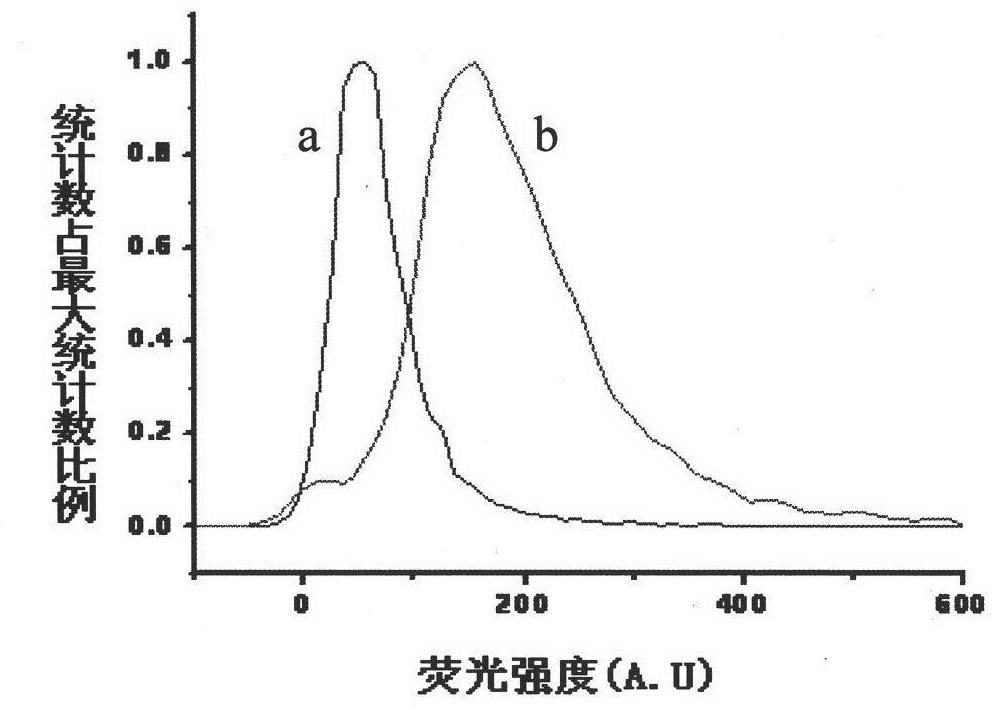
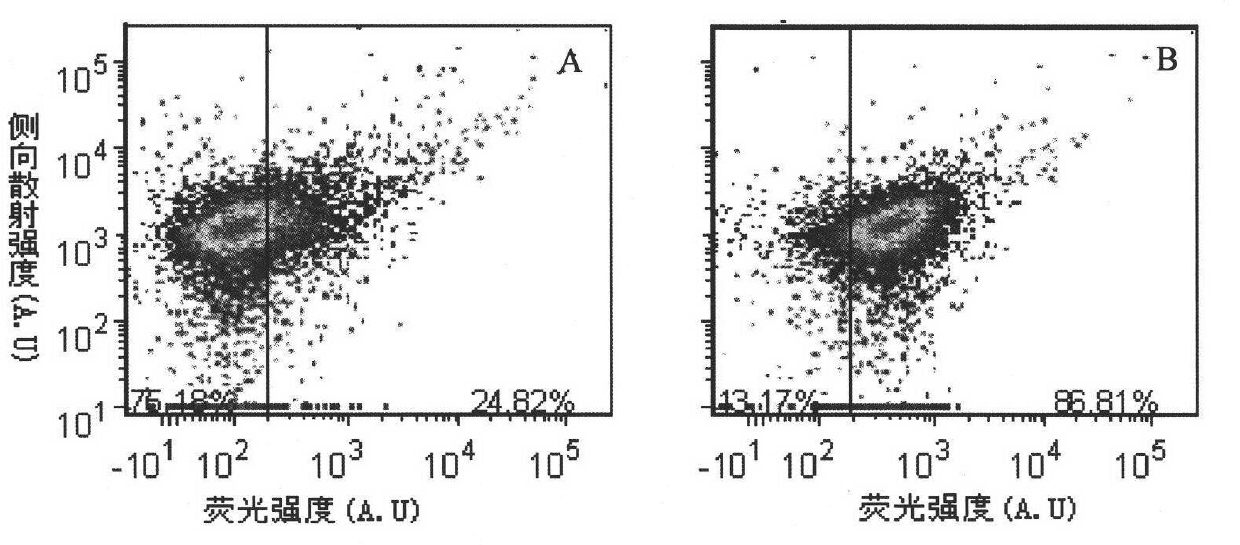
[0084] Pick a single colony of E.coli TG1 to be tested and culture it overnight in 2mL 2×YT, 37°C, 250rpm, take 100L of the overnight cultured E.coliTG1 bacteria solution and add it to 10mL 2×YT-G, shake to OD at 37°C, 250rpm =0.6, take 2 mL of the corresponding bacterial solution in a culture tube, add recombinant phage and prophage respectively, infect at 37° C., 250 rpm for 1 hour. Take 80uL of each infection solution, wash twice with 100L HBSS, finally add HBSS, and add the double arsenic dye FlAsH-EDT with a final concentration of 5μM 2 , incubated for 30 min, centrifuged to discard the supernatant, added detergent BAL, incubated for 15 min, centrifuged to discard the supernatant, repeated washing once, added HBSS 80uL to resuspend. In this way, the bacterial samples infected with the prophage are the negative control group; the bacter...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com