Genetically engineered bacteria for efficiently secreting, expressing and reconstructing cutinase and method for constructing same
A genetically engineered bacteria, secretion and expression technology, applied in the field of bioengineering, can solve the problems of increased host metabolic burden, low expression level of cutinase, etc., and achieve low cost, high recovery rate and obvious effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Extraction of E.coli CFT073 Total DNA
[0036] Cultivate the E.coli CFT073 strain in LB liquid medium (peptone 10g / L, yeast extract 5g / L, NaCl 10g / L) overnight, take 3mL of the bacterial liquid, and centrifuge at 12000rpm for 2min to collect the bacterial cells. Technology Service Co., Ltd. Bacterial Genomic DNA Extraction Kit Method (EZ SpinColumn Bacterial Genomic DNA Isolation Kit UNIQ-10 Column Bacterial Genomic DNA Extraction Kit) to extract the total DNA of E.coli CFT073;
Embodiment 2
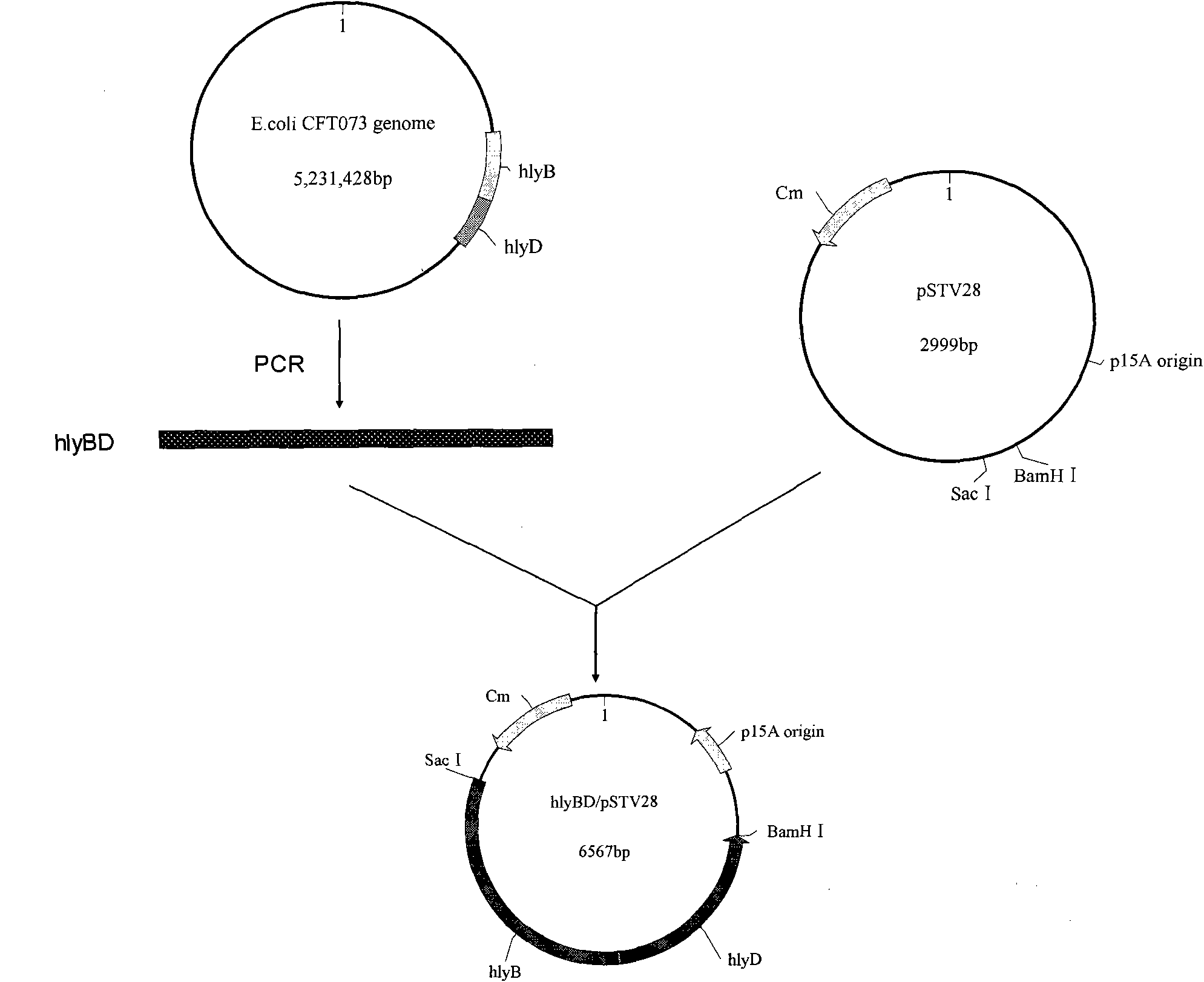
[0037] Embodiment 2: Construction of hlyBD / pSTV28 recombinant plasmid
[0038] Primers P1 and P2 were designed according to the gene sequence of hlyBD.
[0039] P1: 5'-CggCgAgCTCggATTCTTgTCATAAAATTg-3'
[0040] P2: 5'-CCACggATCCTTAACgCTCATgTAAAC-3'
[0041] The hlyBD gene was amplified by PCR using the total DNA of E.coli CFT073 as a template and P1 and P2 as primers. The PCR reaction was carried out in a 50 μL system, and the reaction conditions were 30 cycles of denaturation at 94°C for 1 min, followed by denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 4 min. A PCR fragment of 3579bp was amplified and recovered by tapping the gel. The recovered fragment was ligated with the pMD18-Tsimple vector, the ligated product was transformed into Escherichia coli JM109, and the transformed product was coated on an LB plate containing 30 mg / L chloramphenicol. After culturing overnight at 37°C, single clones were selected, inserted into LB liquid m...
Embodiment 3
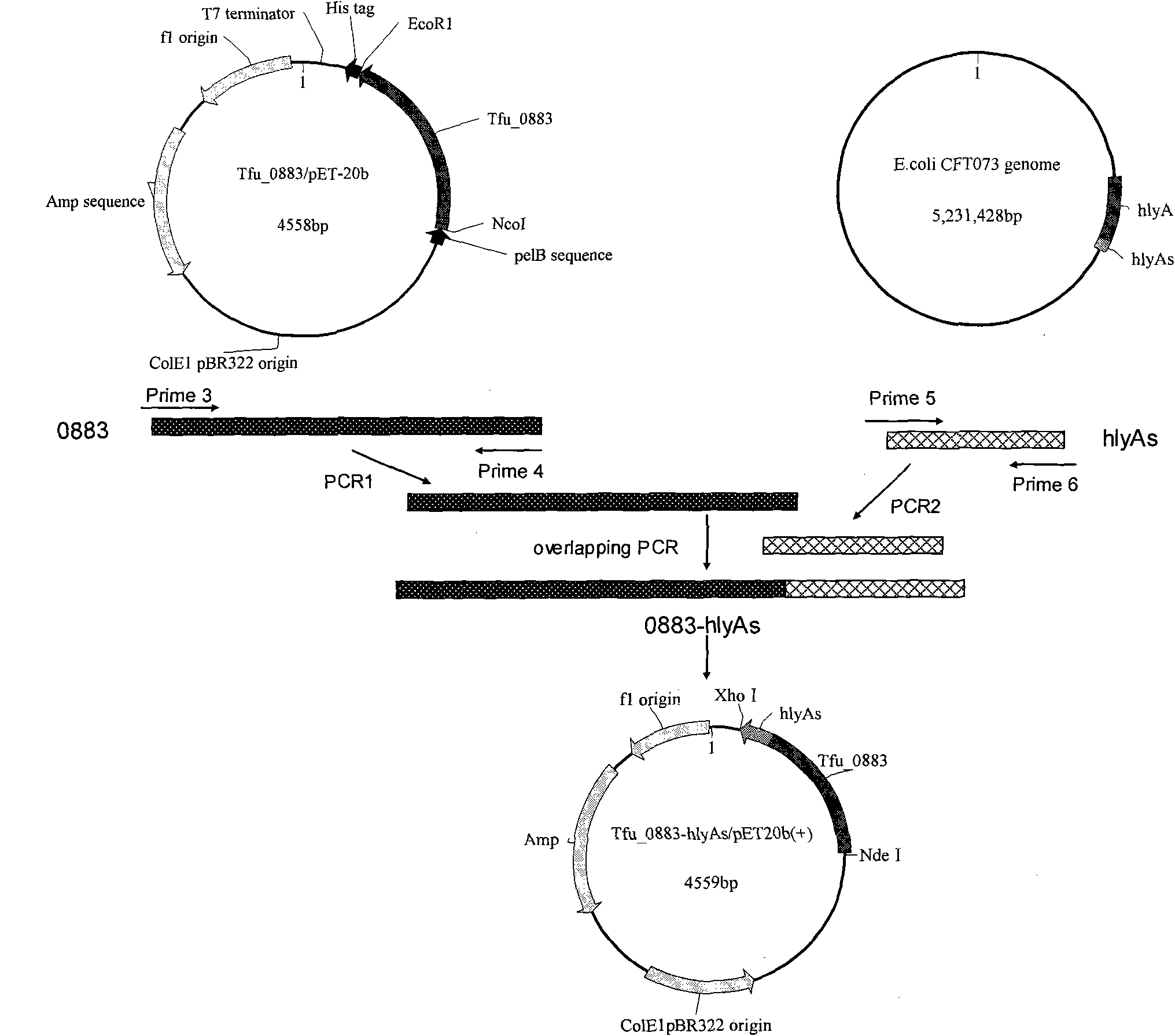
[0105] Embodiment 3: Construction of Tfu_0883-hlyAs / pET20b (+) recombinant plasmid
[0106] Two pairs of primers P3, P4 and P5, P6 were designed according to the sequences of cutinase and hlyAs.
[0107] P3: 5'-gTAATCCATATggCCAACCCCTACgAgCgC-3'
[0108] P4: 5'-gACTTCCATAggCTAAgAACgggCAggTggAg-3'
[0109] P5: 5'-CTCCACCTgCCCgTTCTTAgCCTATggAAgTC-3'
[0110] P6: 5'-CCgCTCgAgTTATgCTgATgCTgTCAAAg-3'
[0111]Using the Tfu_0883-pET20b(+) plasmid DNA constructed earlier in our laboratory as a template and using P3 and P4 as primers, the cutinase gene was amplified by PCR. The hlyAs gene was amplified by PCR using the total DNA of E.coli CFT073 as a template and P5 and P6 as primers. The PCR reaction was carried out in a 50 μL system, and the reaction conditions were denaturation at 94°C for 1 min, followed by a cycle of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min and 20 s, respectively. . PCR fragments of 783bp and 180bp were respec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com