Affinity peptide specific to herceptin and segment thereof
A technology of trastuzumab and affinity peptides, which is applied in the field of affinity peptides, can solve the problems of inability to separate antibodies, harsh elution conditions, and poor chemical stability, and achieve biodegradable, low-cost, and small molecular weight Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: the design of trastuzumab affinity peptide
[0032] The peptide of the present invention is rationally designed based on the complex configuration of the antigen-binding fragment of trastuzumab and HER2 by using molecular dynamics method, and then characterized.
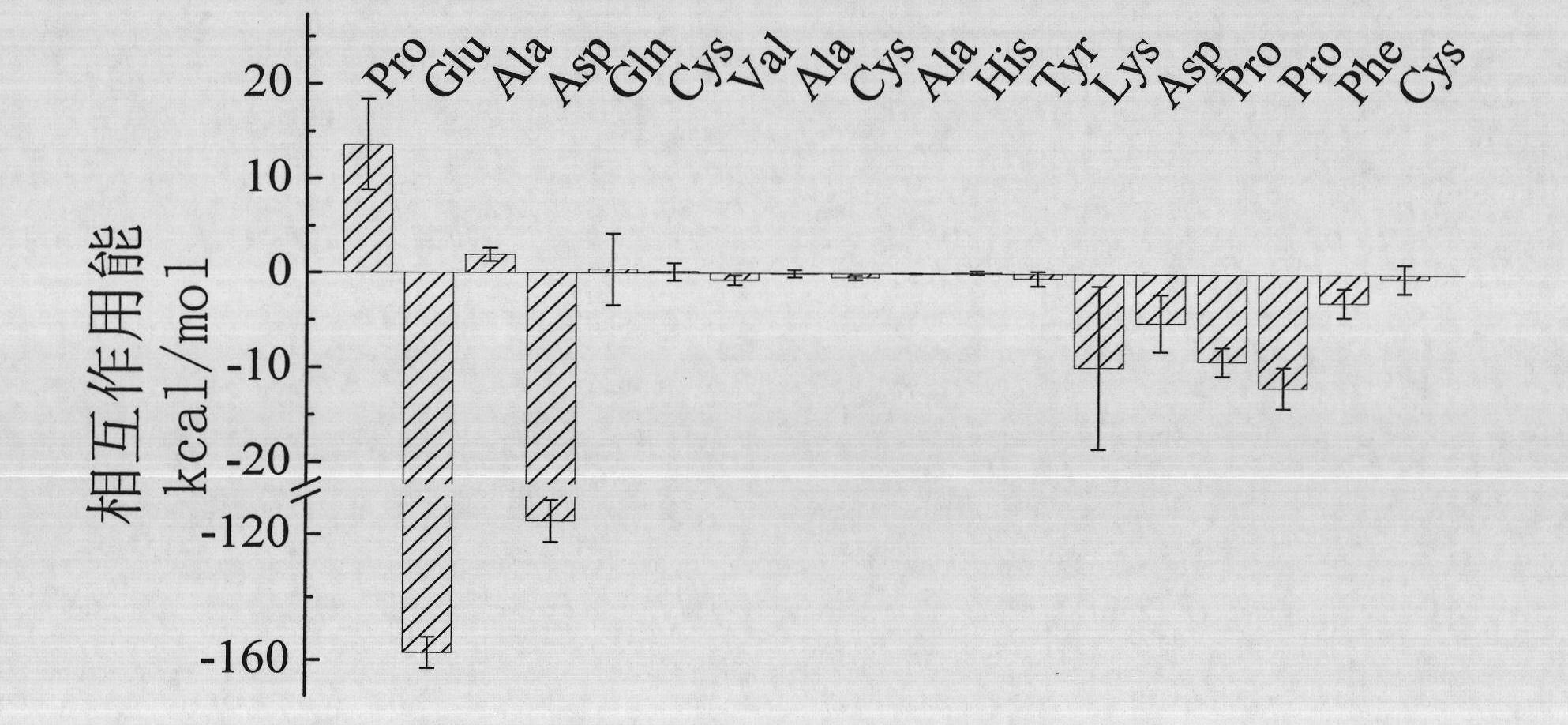
[0033] The three-dimensional structure data (PDB ID: 1N8Z) of the complex of the antigen-binding fragment of trastuzumab and HER2 was determined by Hyun-Soo Cho et al. (Hyun-Soo Cho et al. (2003) Nature, 421: 756-760) . The interaction sites between the two include three loop structures in HER2, loop1: 557-561, loop2: 570-573, and loop3: 593-603. Residues 557-574 of HER2 were intercepted from the structure of the complex to form a structure as follows:
[0034] Pro-Glu-Ala-Asp-Gln-Cys-Val-Ala-Cys-Ala-His-Typ-Lys-Asp-Pro-Pro-Phe-Cys
[0035] The above-mentioned octadecapeptide and the Fab segment of trastuzumab were equilibrated in a water box with periodic boundaries for 4 ns, and the simulati...
Embodiment 2
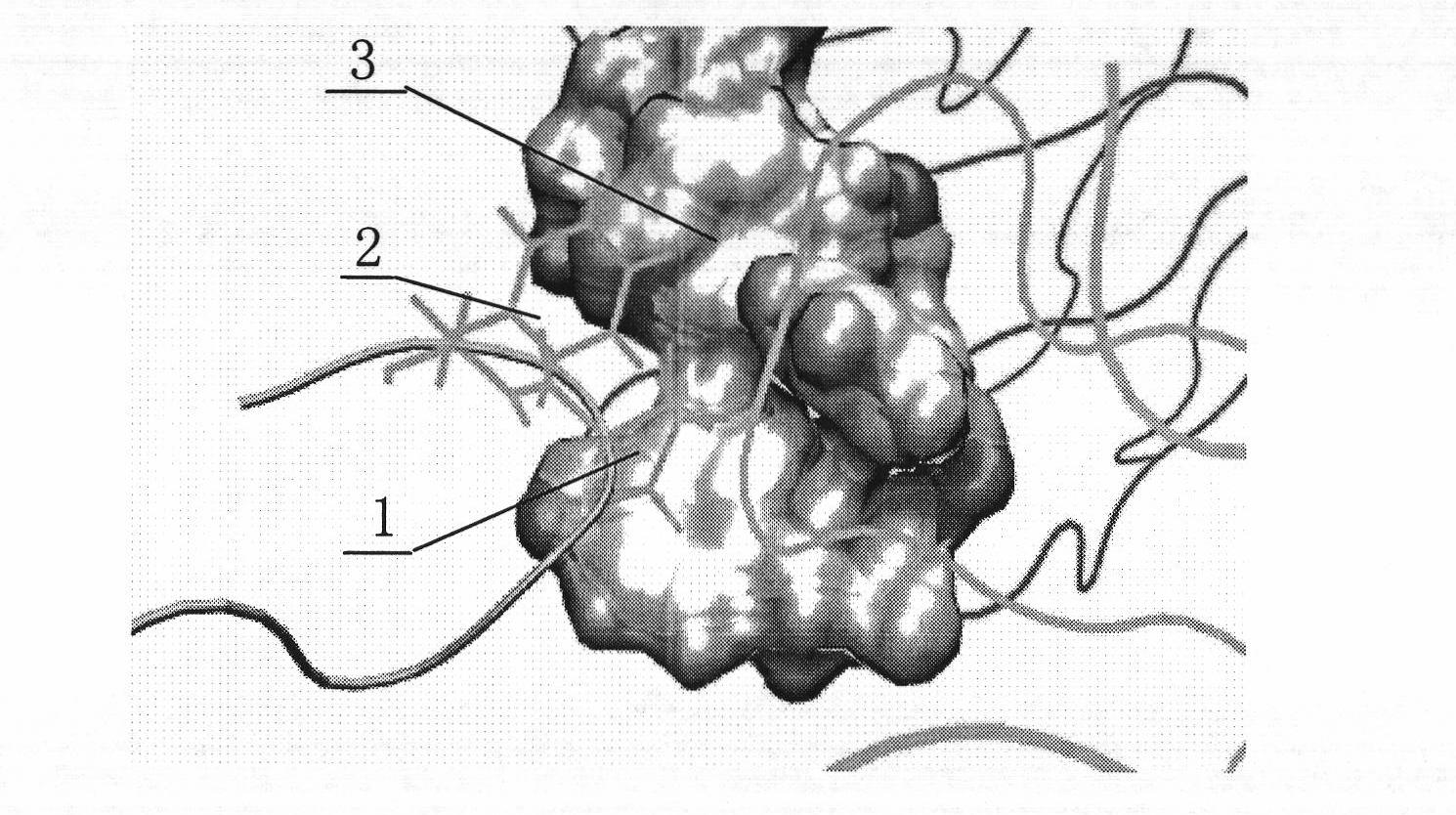
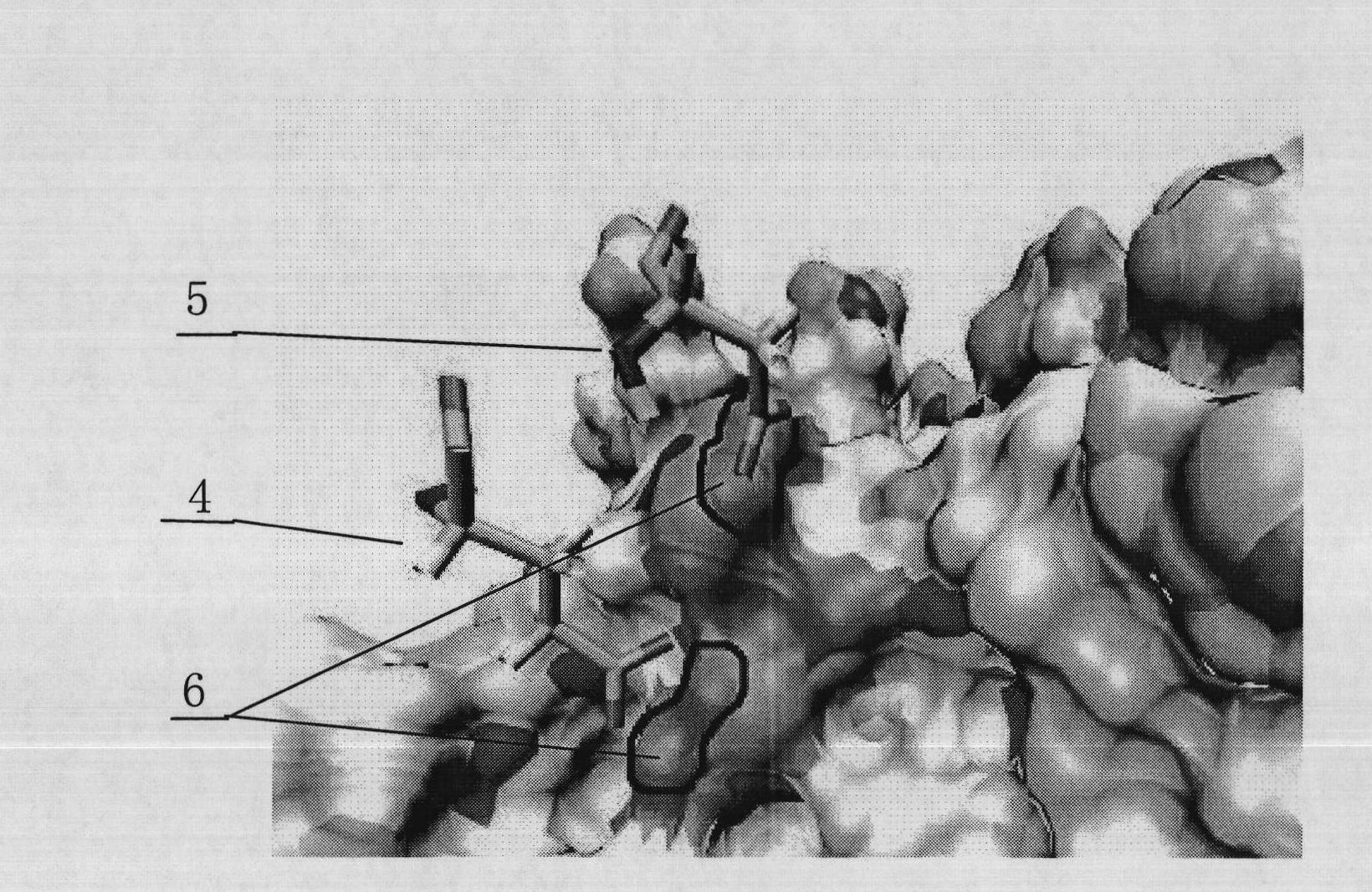
[0045] The affinity peptide specific to the trastuzumab fragment with the structure shown in SEQ ID Nos.11-14 is placed near the binding site on the heavy chain of the trastuzumab Fab segment (residue numbers 33, 50, 55, 59, 99-105), by manipulative molecular dynamics (SMD) method along the centroid of the affinity peptide-trastuzumab Fab segment centroid direction at 0.25 Compress the ligand at a constant speed to make the two tightly combined, and then perform molecular dynamics simulation according to the method and conditions in "Design of Trastuzumab Affinity Peptide", and obtain an optimized affinity peptide-trastuzumab with a relaxation of 4ns The equilibrium configuration of the Fab fragment complex is found by calculating the interaction energy between the two during the relaxation process and reaching the equilibrium state. These structures are specific to the trastuzumab fragment as shown in SEQ ID Nos.8-19 The interaction energy between the affinity peptide and th...
Embodiment 3
[0046] Embodiment 3: the affinity of polypeptide and trastuzumab
[0047] The complete trastuzumab was constructed, the PDB ID of its FC segment was 3D6G, and the amino acid sequence of the humanized part (greater than 95%) of the complete trastuzumab came from Taxonomy ID 9606. Investigate the affinity peptide and trastuzumab specific for the trastuzumab fragment as shown in SEQ ID Nos.14 in the above-mentioned preference according to the method in "the affinity of the polypeptide and the Fab fragment of trastuzumab" Anti-affinity, its interaction energy size is 210kJ / mol.
[0048] The above shows that the affinity peptide specific to trastuzumab fragments obtained through rational design and interaction energy screening of the present invention can be well combined with trastuzumab or its fragments, using this affinity The purpose of separating, purifying or detecting it can be achieved.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com