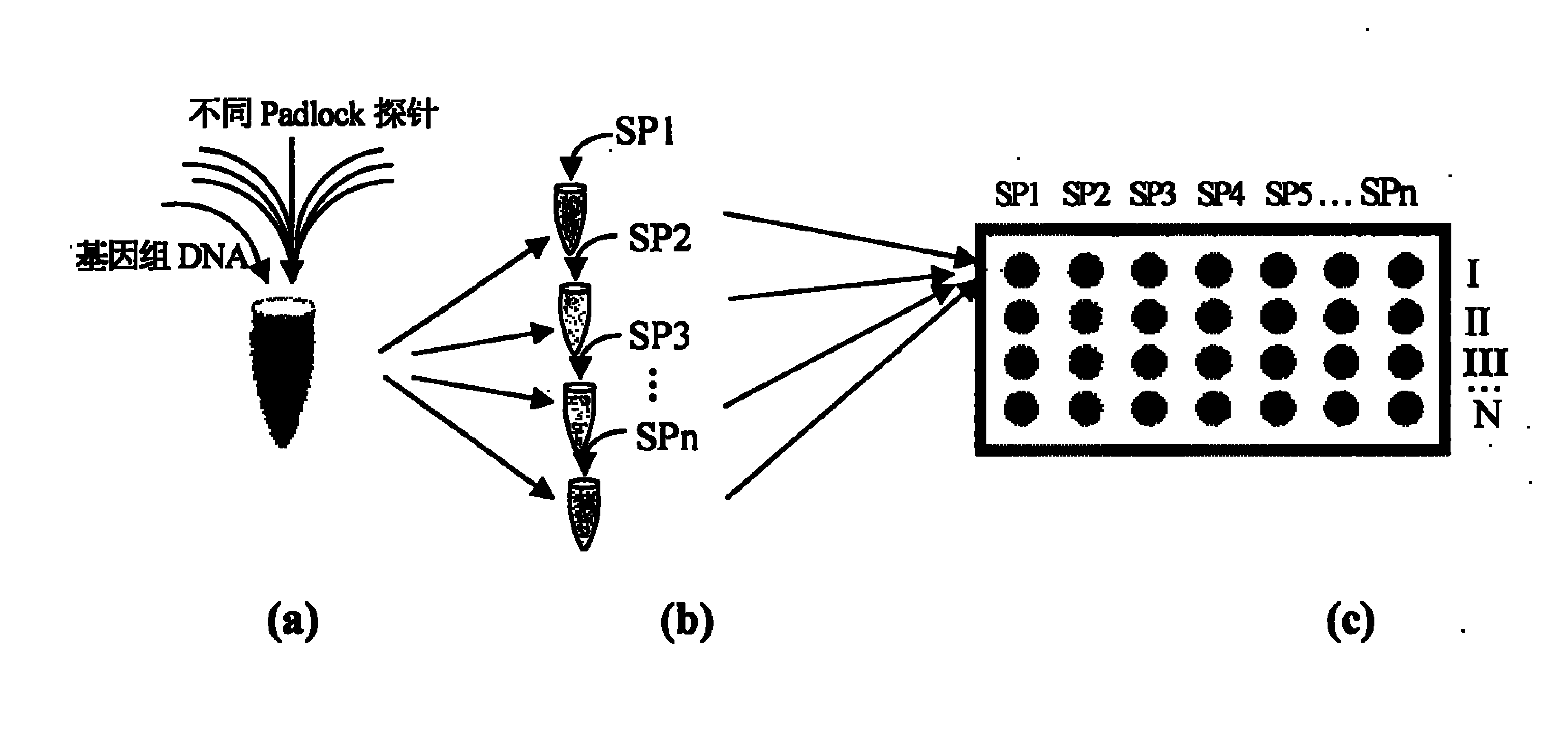
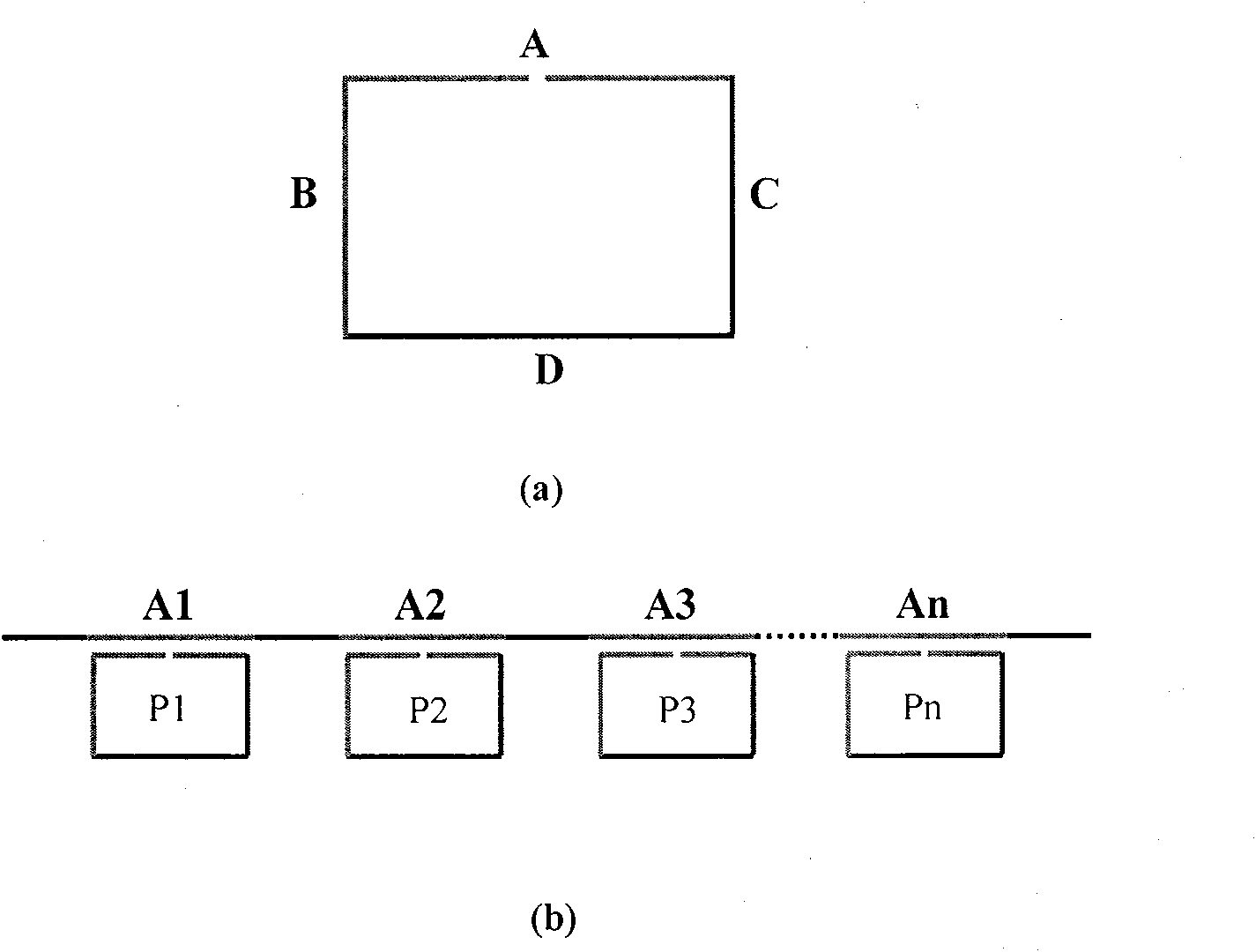
Multi-sample multi-site SNP detection method
A detection method and multi-site technology, applied in fluorescence/phosphorescence, material excitation analysis, etc., can solve problems such as the difficulty in optimizing the temperature difference extension conditions of the melting chain, the difficulty in multi-site SNP detection, and the difficulty in micro-sample detection, etc., to achieve Low cost, high sensitivity and specificity, high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Realized the correlation study between 10 polymorphic sites of COMT gene (rs15761987, rs15761985, rs15761983, rs15761981, rs15761980, rs15761978, rs15761977, rs15761976, rs15761975, rs15761967) and postoperative fentanyl analgesic dose.
[0022] For the above 10 SNP sites, design corresponding Padlock probes (18bp at both ends: respectively complementary to the sequences on both sides of the SNP site, and the SNP site corresponds to the base at the 5' end; the middle length is 50bp (see attached table) 1(1)); specific amplification primers with acrylamide modification at the 5' end (complementary to the 18 nucleotide sequences at the 5' end of the Padlock probe) and general fluorescent detection probes (see attached table 1(2) for Detect SNP mutant sites; see attached table 1 (3) for detecting SNP wild-type sites); use different Padlock probes to capture 10 SNP sites of the same sample in the same reaction tube, and connect the Padlock probes with ligase Needle, the Pad...
Embodiment 2
[0024] Detection of 4 SNP sites (rs6929137(G / A), rs1999805(T / C), rs4870044(G / A), rs1038304(A / G)) in the chromosome 6q25.1 region of 200 people, and further research Association of gene polymorphisms with bone mineral density and vertebral fractures in postmenopausal women
[0025]For the four SNP sites of rs6929137(G / A), rs1999805(T / C), rs4870044(G / A), rs1038304(A / G), corresponding Padlock probes, specific amplification primers and universal fluorescence detection were designed Probes (sequence features are all the same as in Example 1); in the same reaction tube, use different Padlock probes to capture four SNP sites of the same sample, and connect the Padlock probes with ligase, and there are Padlock probes corresponding to the SNP sites Circularization occurs, otherwise no circularization; then use specific amplification primers to hybridize with Padlock probes and perform constant temperature rolling circle amplification with high-fidelity Bst, circularized Padlock probes ...
Embodiment 3
[0027] Realize the correlation between the pain degree of patients with rheumatoid arthritis and the SNP of HLA-II gene
[0028] For 3 SNP sites (rs1059576, rs1059582, rs1059586) of HLA-DRB1 gene, 2 SNP sites (rs2071282, rs2071283) of HLA-NOTCH4 gene and 1 SNP site (rs1049110) of HLA-DQB2 gene, design corresponding Padlock probes (18bp at both ends and 50bp in the middle), specific amplification primers and universal fluorescent detection probes with biotin modification at the 5' end (sequence features are the same as in Example 1); in the same reaction tube Use different Padlock probes to capture 6 SNP sites of the same sample, and connect the Padlock probes with ligase, and the Padlock probes with corresponding SNP sites will be circularized, otherwise they will not be circularized; then use biotin-modified Specific amplification primers were hybridized with Padlock probes, and high-fidelity Bst was used for constant temperature rolling circle amplification. The circularized...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com