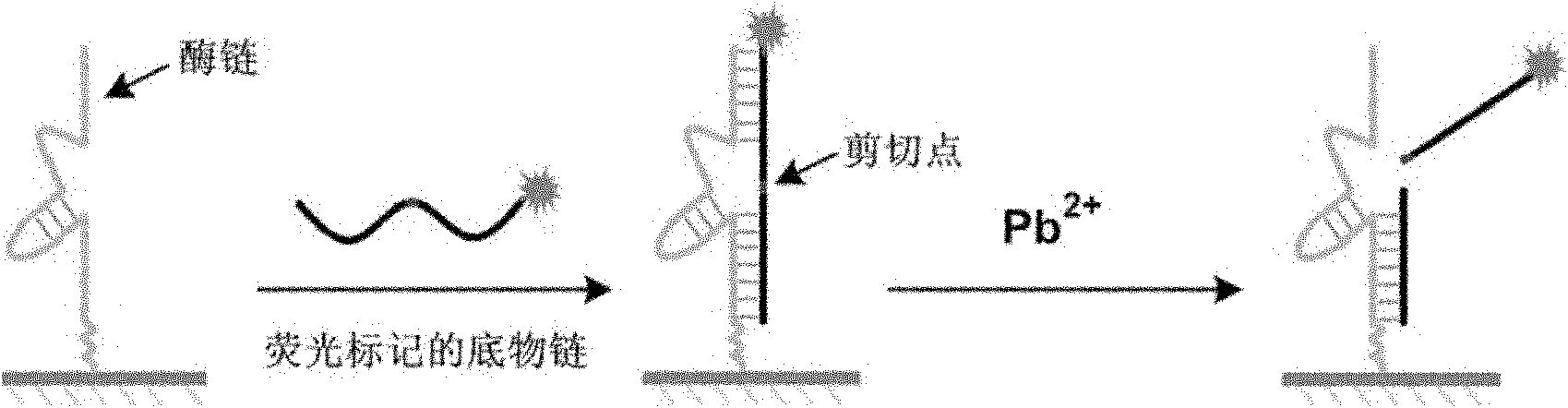
Lead ion detection chip based on deoxyribonuclease as well as making and application methods
A deoxynuclease and detection chip technology, applied in the field of biological analysis, can solve the problems of cumbersome operation and unsuitability for high-throughput detection, and achieve the advantages of simple production method, low reagent consumption, high detection sensitivity and specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1: utilize probe Pb-1 and Pb-2 to prepare Pb 2+ Detection chip.
[0027] Dissolve 40 μM Pb-2 in water, then mix it with the same volume of Spotting Solution, use Cartesian’s microarray chip production system to array on the surface of the aldehyde-modified glass slide, and place it at room temperature at 70% relative humidity Store for 48-72 hours at room temperature for fixation, then immerse the slide in 0.2% SDS at room temperature for a few minutes, then immerse in pure water for a few minutes, then immerse in 0.2% SDS twice, each time for 2 minutes, and then immerse in pure water Twice, 2min each time, let dry. Pb-1 was diluted with hybridization solution (10mM Tris-HCl, pH 7.2, 1M NaCl) to a final concentration of 5 μM, dropped on the chip, covered with a cover slip, and hybridized at room temperature for 12-16 hours. Then put the chip at 4°C for 30min, then at 25°C for 15min. Then wash with 0.2% SDS, 2×SSC, 0.2×SSC for 3 minutes, and dry it for late...
Embodiment 2
[0028] Embodiment 2: utilize probe Pb-3 and Pb-4 to prepare Pb 2+ Detection chip.
[0029] Dissolve 40 μM Pb-4 in water, then mix it with the same volume of Spotting Solution, use Cartesian’s microarray chip production system to array on the surface of aldehyde-modified glass slides, store at room temperature and 70% relative humidity Fix for 48-72 hours, then immerse the slide in 0.2% SDS at room temperature for a few minutes, then immerse in pure water for a few minutes, then immerse in 0.2% SDS twice, each time for 2 minutes, then immerse in pure water twice , each 2min, dry. Dilute Pb-3 with hybridization solution (10mM Tris-HCl, pH 7.2, 100mM NaCl) to a final concentration of 5μM, drop on the chip, cover with a cover glass, and hybridize at room temperature for 12-16h, then place the chip at 4°C, 30min, then 15min at 25°C. Then wash with 0.2% SDS, 2×SSC, 0.2×SSC for 3 minutes, and dry it for later use.
Embodiment 3
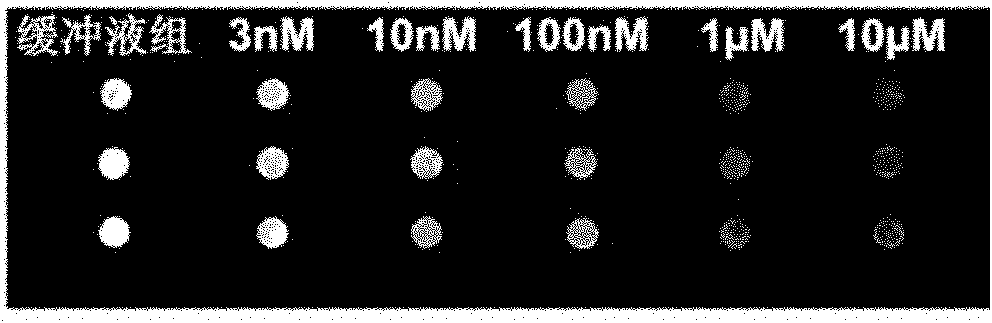
[0030] Embodiment 3: Using the chip prepared by probe Pb-1 and Pb-2 to detect different concentrations of Pb 2+ .
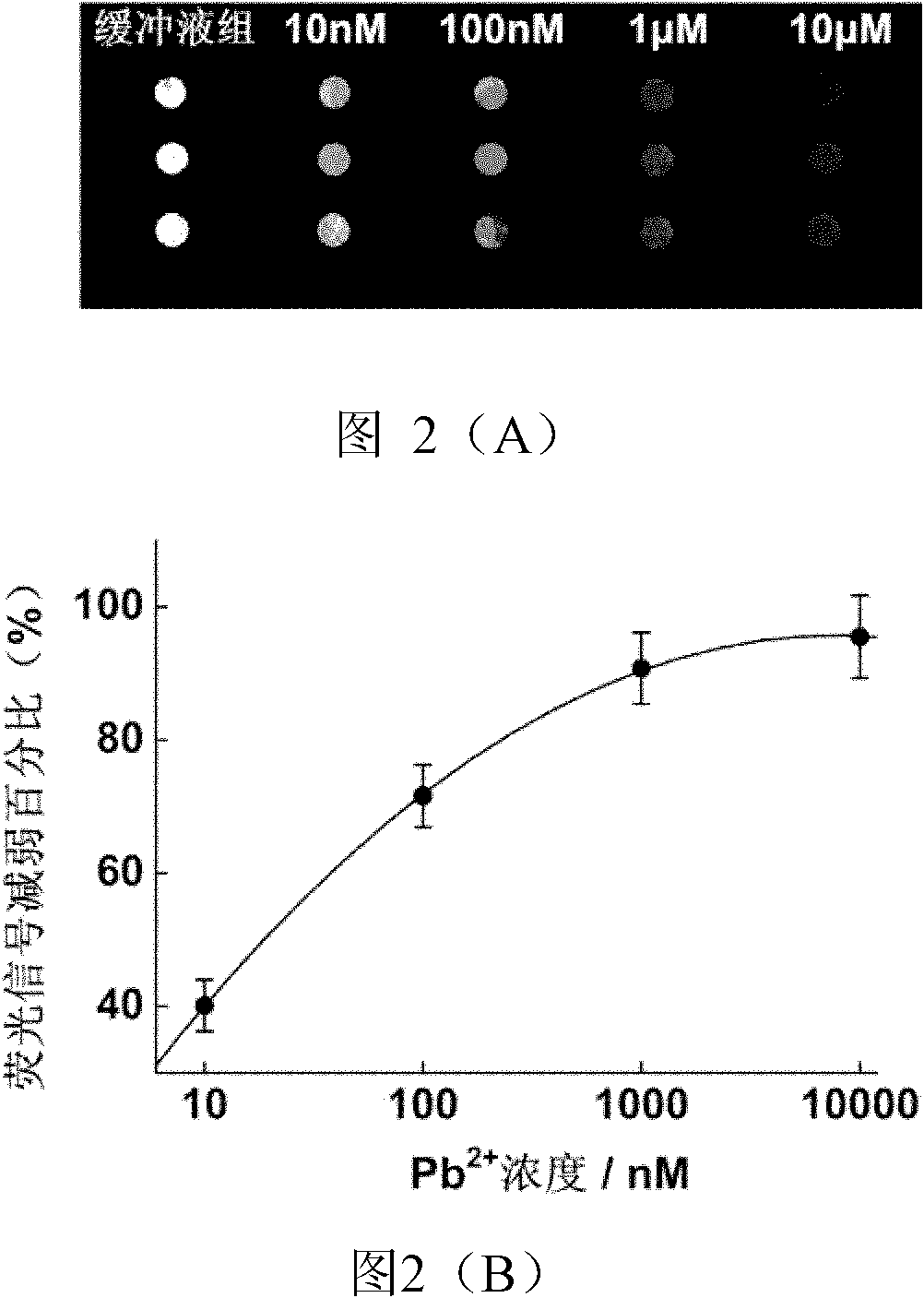
[0031] Prepare different concentrations of Pb by diluting with 10mM Tris-HCl, pH 7.2, 50mM NaCl 2+ , with different concentrations of Pb 2+ The solution was placed on the prepared chip spots at 4°C for 1 hour, and then left at room temperature for 15 minutes. And washed 3 times with buffer, and dried. Scan the photos with the chip signal analysis system Scanarray 3000 of General Scanning Company ( figure 2 A) and analyze the results ( figure 2 B).
[0032] The results showed that in Pb 2+ In the presence of ions, the fluorescence intensity at the chip spot weakens, when Pb 2+ When the concentration was 10nM, compared with the fluorescence intensity of the buffer group, the fluorescence intensity at the spot was weakened by 40%. 2+ As the concentration increases, the fluorescence signal gradually decreases. The chip detects Pb 2+ The limit of detection...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com