Efficient gene knockout method of pseudomonas putida KT2440
A Pseudomonas putida, gene knockout technology, applied in the field of genetic engineering, can solve the problems of difficult high-quantification of vectors, increased cost, labor consumption, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1. Construction of a plasmid for the expression of recombinase induced by m-toluic acid
[0064] 1. Preparation of Escherichia coli DH10B electroporation competent and DNA transformation
[0065] Inoculate a freshly streaked single colony from the plate into 2ml LB liquid medium 37 o C Shake overnight, transfer to 50 ml LB at 1 / 50 volume, shake culture to the OD of the bacteria 600 About 0.6. Pour the bacterial solution into a pre-cooled centrifuge tube and place in an ice bath for 10 minutes, 4 o C, centrifuge at 5000rpm for 5 minutes, discard the supernatant. Wash the precipitate twice with 10% glycerol, and finally suspend in 200ml of 10% glycerol, and divide into 50ml tubes.
[0066] Add the DNA dissolved in TE buffer or double distilled water to 50ml DH10B electroporation competent cells melted on ice, flick and mix well. The mixture was transferred to a pre-cooled 1mm electroporation cuvette on ice, and transformed by electric shock. Electroporation ...
Embodiment 2
[0085] Example 2. Knockout of 1.0 kb fragment of Pseudomonas putida KT2440 genome PP_3948 gene
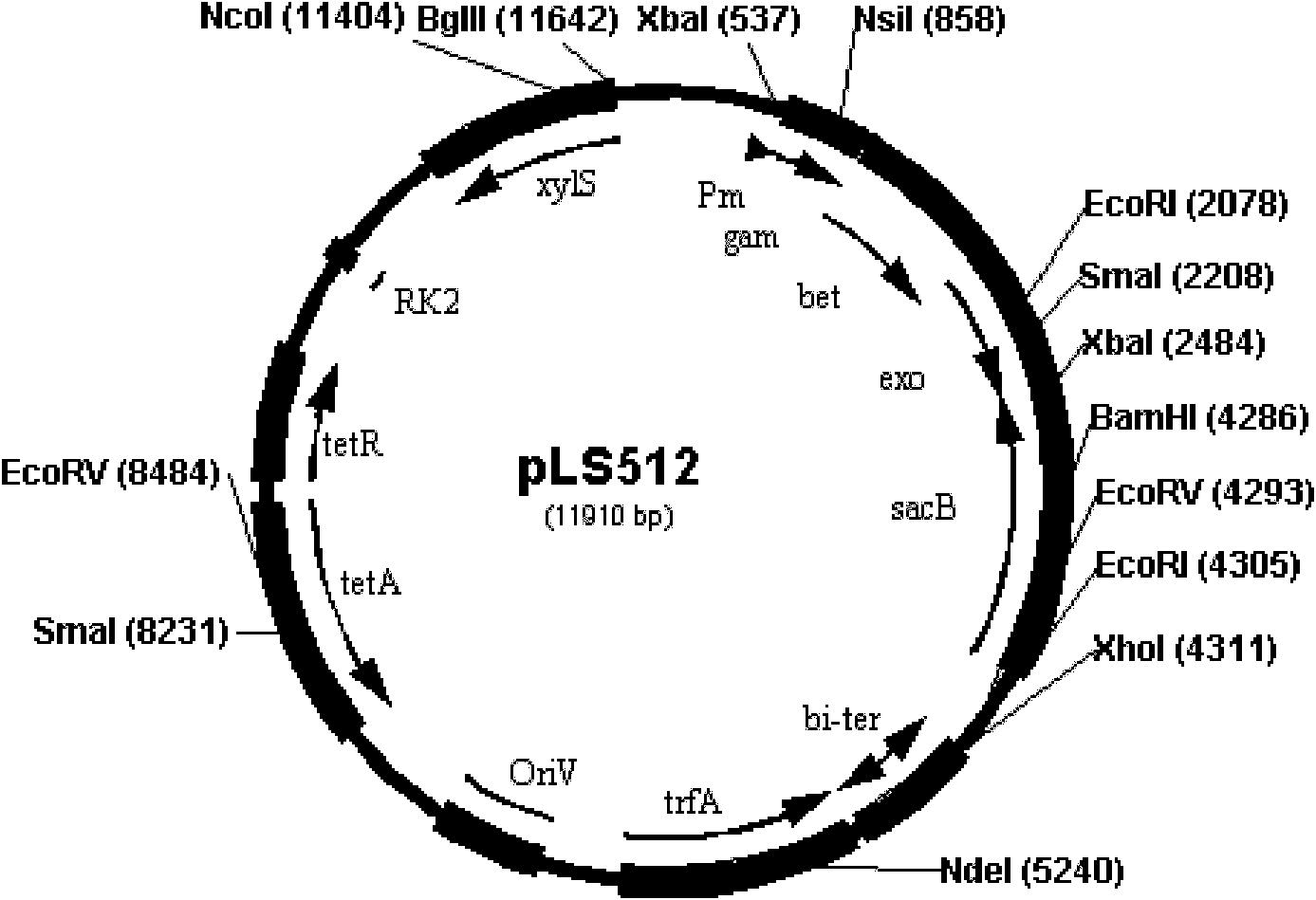
[0086] 1. Preparation of Pseudomonas putida KT2440 electroporation competent cells and transformation of pLS512
[0087] from storage at -70 o Streak the Pseudomonas putida KT2440 cryopreservation tube of C into LB solid medium, culture overnight, pick a single colony and put it into 2ml EM liquid medium for 30 o C Shake overnight, transfer to 50 ml EM medium at 1 / 50 volume, and shake to culture the cells to OD 600 About 0.6. Pour the bacterial solution into a pre-cooled centrifuge tube and place in an ice bath for 10 minutes, 4 o C, centrifuge at 7000rpm for 5 minutes, discard the supernatant. The cells were washed 3 times with ice-cold 3 mM 4-hydroxyethylpiperazineethanesulfonic acid (HEPES, pH=7.0), the cells were concentrated 300 times, aliquoted in 50 ml and used for one electrotransformation.
[0088] Transformation of pLS512: Add pLS512 dissolved in TE buffer to 50ml ...
Embodiment 3
[0103] Example 3. Knockout of PP_1384 to PP_1388 7.3kb fragment of Pseudomonas putida KT2440 genome
[0104] The preparation and DNA transformation of the electrotransformation competent cells of Pseudomonas putida KT2440 / pLS512 induced to express the recombinant enzyme were the same as in Example 2.
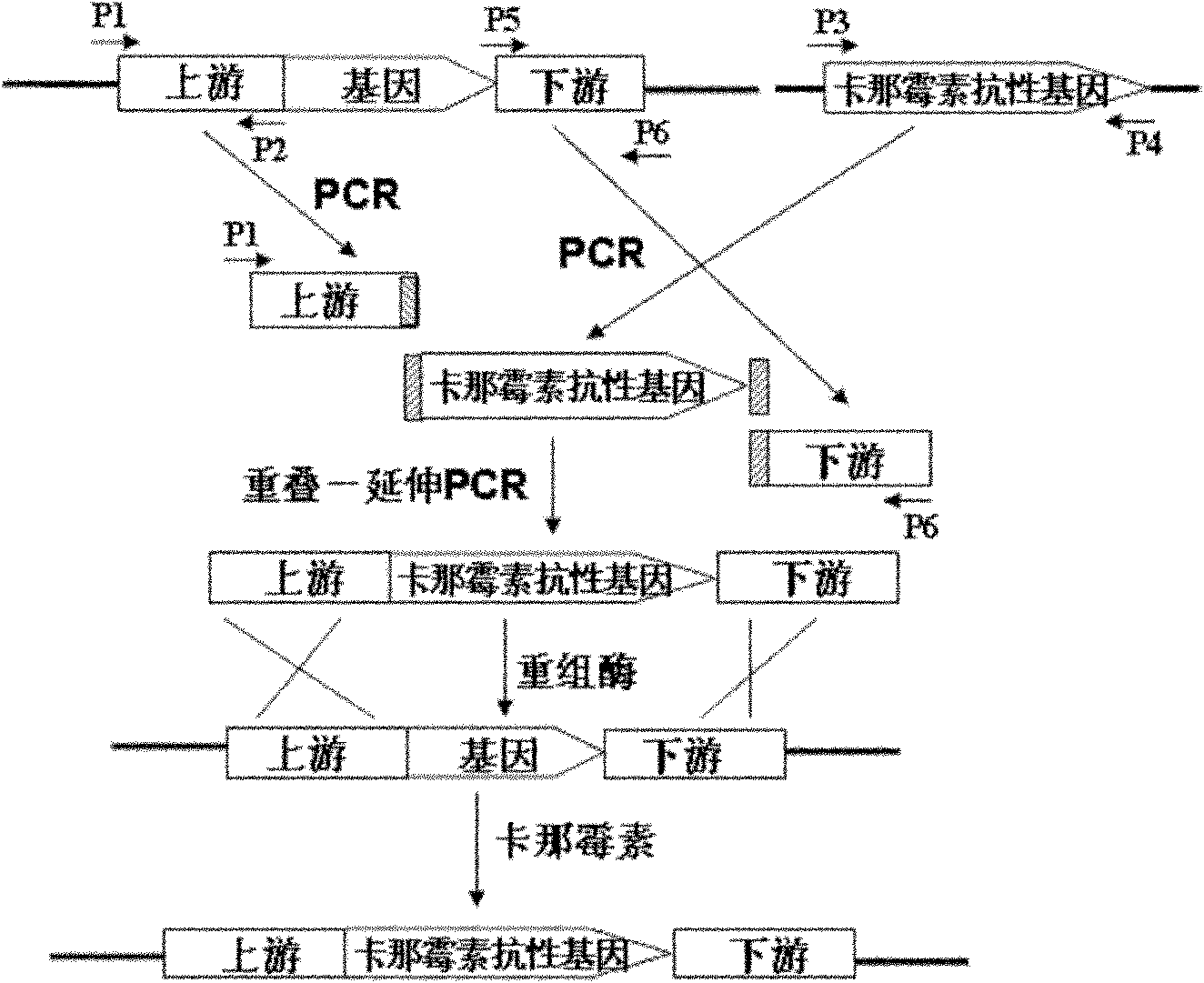
[0105] Primers were designed to amplify the kanamycin resistance gene containing homologous fragments by PCR. KSD1: 5'-GTCGACGA AGTTGCGGTTGATG, (SEQ ID NO. 13); KSD2: 5'-CCGGTTCGCTTGCTGTC CATATGATCGGCCTTGTTGCAAAAGC, (SEQ ID NO. 14); KSD3: 5'-GCTTTT GCAACAAGGCCGATCATATGGACAGCAAGCGAACCGG, (SEQ ID NO. 15); KSD4: 5'- GAC AAAACAAGGTATTCCCGTGTCAGAAGAACTCGTCAAGAA G, (SEQ ID NO. 16); KSD5: 5'-CTTCTTGACGAGTTTCTTCTGACACGGGAATA CCTTGTTTTGTC, (SEQ ID NO. 17); KSD6: 5'-GCGTGCCGACATTGGGTCG AAC, (SEQ ID NO. 18). There are 43 base pairs of overlap between KSD2 and KSD3 and between KSD4 and KSD5, and they are all presented in the form of reverse complementarity.
[0106] Using the genomic DNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com