Nosiheptide resistance ribosomal RNA methyltransgerase, and preparation method and application thereof
A methyltransferase and ribosome technology, applied in the biological field, can solve problems such as antibiotic failure and bacterial drug resistance, and achieve the effect of reducing drug resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Discovery of nosiheptide-resistant RNA methyltransferase
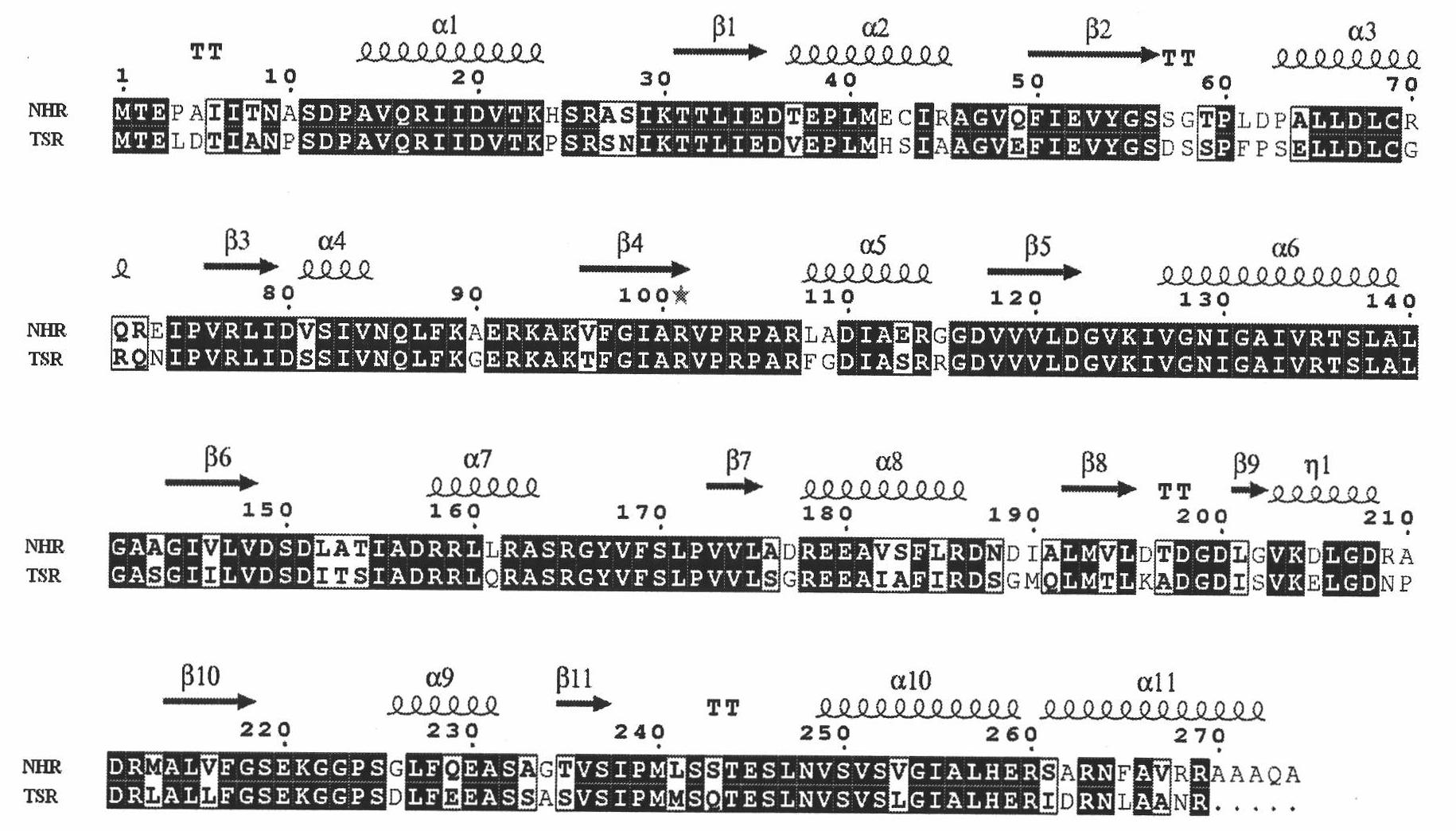
[0065] According to the amino acid sequence of the homologous protein thiostrepton drug-resistant RNA methyltransferase, a Blast comparison was performed on the website of the European Center for Biology, and the results showed that there was a highly conserved homology in Streptomyces actuosus protein, accession number AAB17875. Previous studies have shown that the fragment containing this protein can endow the bacteria Streptomyces lividans 1326 with resistance to nosiheptide antibiotics. The present invention named it Nosiheptide Resistance RNA Methyltranferase (NHR). The full-length NHR is a protein containing 274 amino acids with a molecular weight of about 29Kd. It is divided into two parts, N-terminal and C-terminal. domain. Among them, the N-terminal contains 4 α-helices and 4 β-sheets, and forms a sandwich structure responsible for recognizing and binding substrate RNA. The C-terminus cont...
Embodiment 2
[0066] Example 2: Cloning of nosiheptide-resistant RNA methyltransferase gene and construction of prokaryotic expression plasmid pET28A-NHR
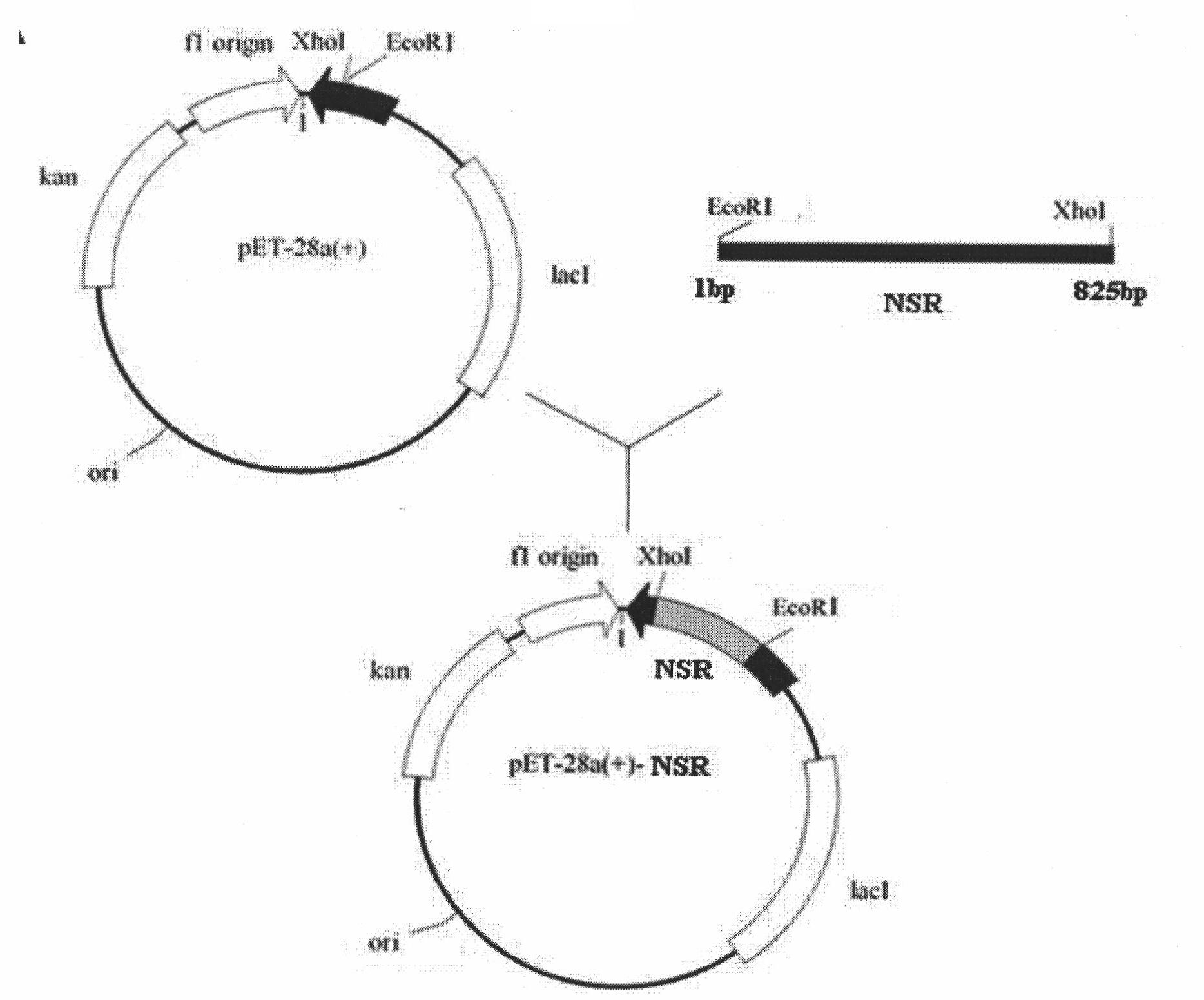
[0067] According to the coding sequence of the nosiheptide drug-resistant RNA methyltransferase gene, primers were designed, and PfuDNA polymerase was used to amplify the nosiheptide drug-resistant RNA methyltransferase gene fragment, which was ligated into The pET28A plasmid forms the pET28A-NHR recombinant plasmid, which is transformed into Escherichia coli BL21, the positive clones are screened by enzymatic hydrolysis, and the nucleotide sequence analysis confirms that the gene sequence is the same as the design. It was confirmed that positive clones were obtained. figure 2 The construction procedure of the pET28A-NHR vector is shown.
[0068] Restriction endonucleases were purchased from Takara Company, Escherichia coli BL21, and plasmid pET28A was preserved in our laboratory.
Embodiment 3
[0069] Embodiment 3: the prokaryotic expression of NHR gene
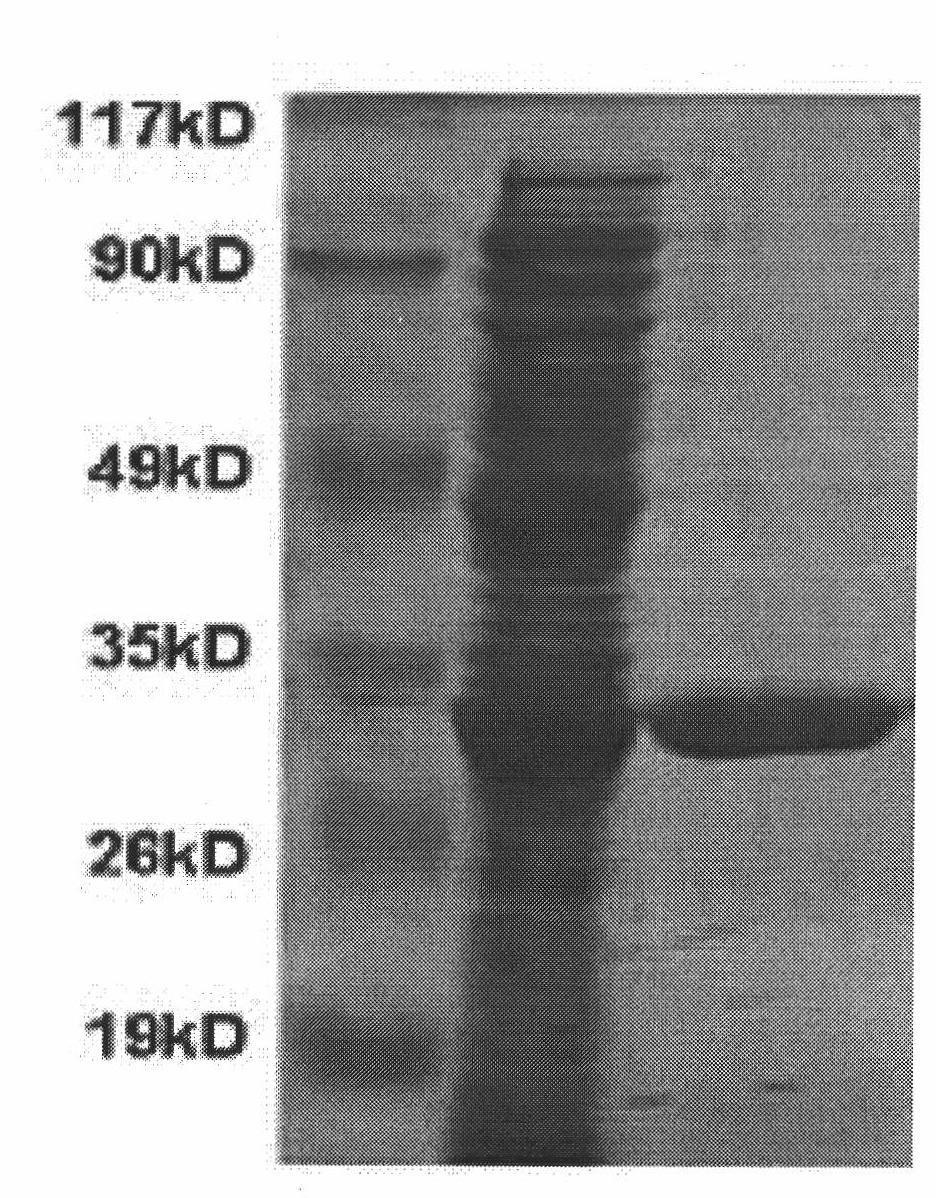
[0070] Pick several positive clonal colonies, shake culture overnight at 30°C in LB medium, then dilute with new LB medium 1:50, shake culture at 30°C until OD600=0.5, add final concentration of 1mmol of IPTG, Expression was induced for 4 hours. After the induction ended, the bacteria were collected by centrifugation, washed twice with PBS (pH=7.4), and the expression product was analyzed by SDS-PAGE. After staining with Coomassie Brilliant Blue R-250, it could be seen that the supernatant after induction was at a molecular weight of 30Kd (considering that the protein There are 6 histidine tags at the end, so the molecular weight of the expressed product is slightly higher).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com