Fast detection method for clostridium perfringens, detection primer group and detection kit
A technology of Clostridium perfringens and detection primers, which is applied in the field of microbial detection to achieve accurate detection, reduce detection links, and shorten the detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Clostridium perfringens PCR detection
[0027] In this embodiment, irrigation water is used as a detection sample, and the specific detection steps are:
[0028] 1. Extraction of sample DNA
[0029] 1.1. Take 1mL of the water sample to be tested and put it into 9mL blister liquid culture medium, and apply the Mart II anaerobic system to incubate for 24 hours at 36°C±1°C after sealing.
[0030] 1.2. Use bacterial genome DNA extraction kit to extract bacterial DNA from cultured products.
[0031] 2. PCR reaction
[0032] 2.1. The upstream and downstream primers of the artificially synthesized Clostridium perfringens outer primers, wherein the upstream primer F3 has the nucleotide sequence shown in SEQ No.1, and the downstream primer B3 has the nucleotide sequence shown in SEQ No.2 acid sequence.
[0033] 2.2. PCR reaction system
[0034] The PCR reaction system is: DNA polymerase 0.05U / μL, dATP, dTTP, dGTP, dCTP each 0.2mM, MgSO 4 2 mM, 10% (volume) 10×DN...
Embodiment 2
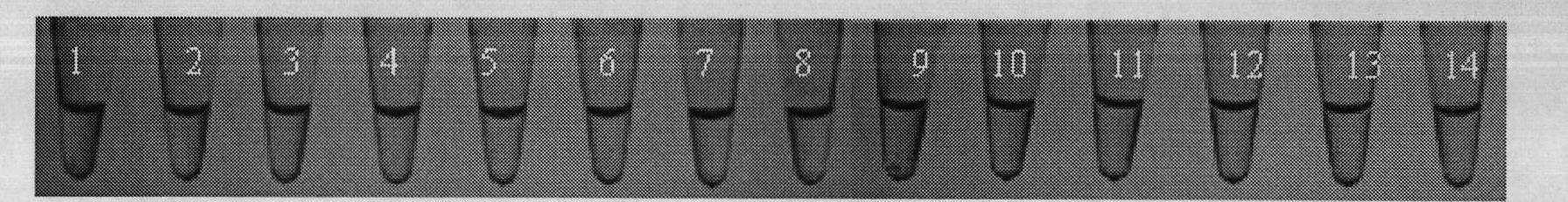
[0041] Example 2 Clostridium perfringens LAMP rapid detection of Mg 2+ concentration test
[0042]In this embodiment, irrigation water is used as a detection sample, and the specific detection steps are:
[0043] 1. Extraction of sample DNA
[0044] 1.1. Take 1mL of the water sample to be tested and put it into 9mL blister liquid culture medium, and apply MartⅡ anaerobic system after sealing for 24 hours at 36°C±1°C.
[0045] 1.2. Use bacterial genome DNA extraction kit to extract bacterial DNA from cultured products.
[0046] 2. LAMP rapid detection
[0047] 2.1. Artificially synthesized Clostridium perfringens LAMP detection primer set, the upstream primer F3 of the outer primer has the nucleotide sequence shown in SEQ No.1; the downstream primer B3 of the outer primer has the sequence shown in SEQ No.2 The nucleotide sequence shown; the upstream primer FIP of the inner primer has the nucleotide sequence shown in SEQ No.3; the downstream primer BIP of the inner primer ha...
Embodiment 3
[0057] Example 3 Clostridium perfringens LAMP rapid detection sensitivity test
[0058] In this embodiment, irrigation water is used as a detection sample, and the specific detection steps are:
[0059] 1. Extraction of sample DNA
[0060] 1.1. Take 1mL of the water sample to be tested and put it into 9mL blister liquid culture medium, and apply the Mart II anaerobic system to incubate for 24 hours at 36°C±1°C after sealing.
[0061] 1.2. Use bacterial genome DNA extraction kit to extract bacterial DNA from cultured products.
[0062] 2. LAMP rapid detection
[0063] 2.1. Artificially synthesized Clostridium perfringens LAMP detection primer set, the upstream primer F3 of the outer primer has the nucleotide sequence shown in SEQ No.1; the downstream primer B3 of the outer primer has the sequence shown in SEQ No.2 The nucleotide sequence shown; the upstream primer FIP of the inner primer has the nucleotide sequence shown in SEQ No.3; the downstream primer BIP of the inner pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com