nfi-gene-knocked-out mutant strain of escherichia coli DH5 alpha as well as preparation method and application thereof
A technology of Escherichia coli and gene knockout, applied in the application field of sequencing or/and cloning research, to achieve the effect of stabilizing biological properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
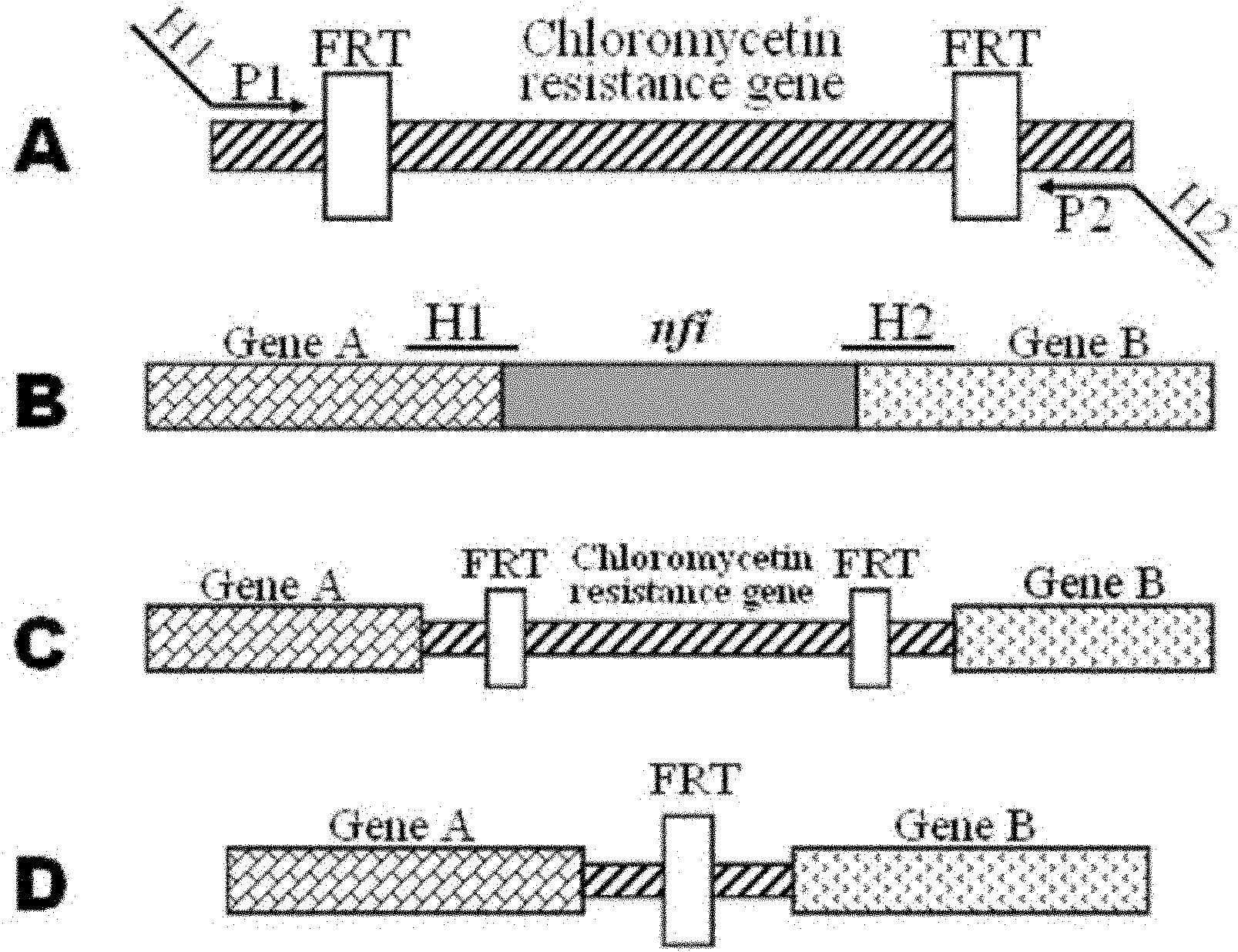
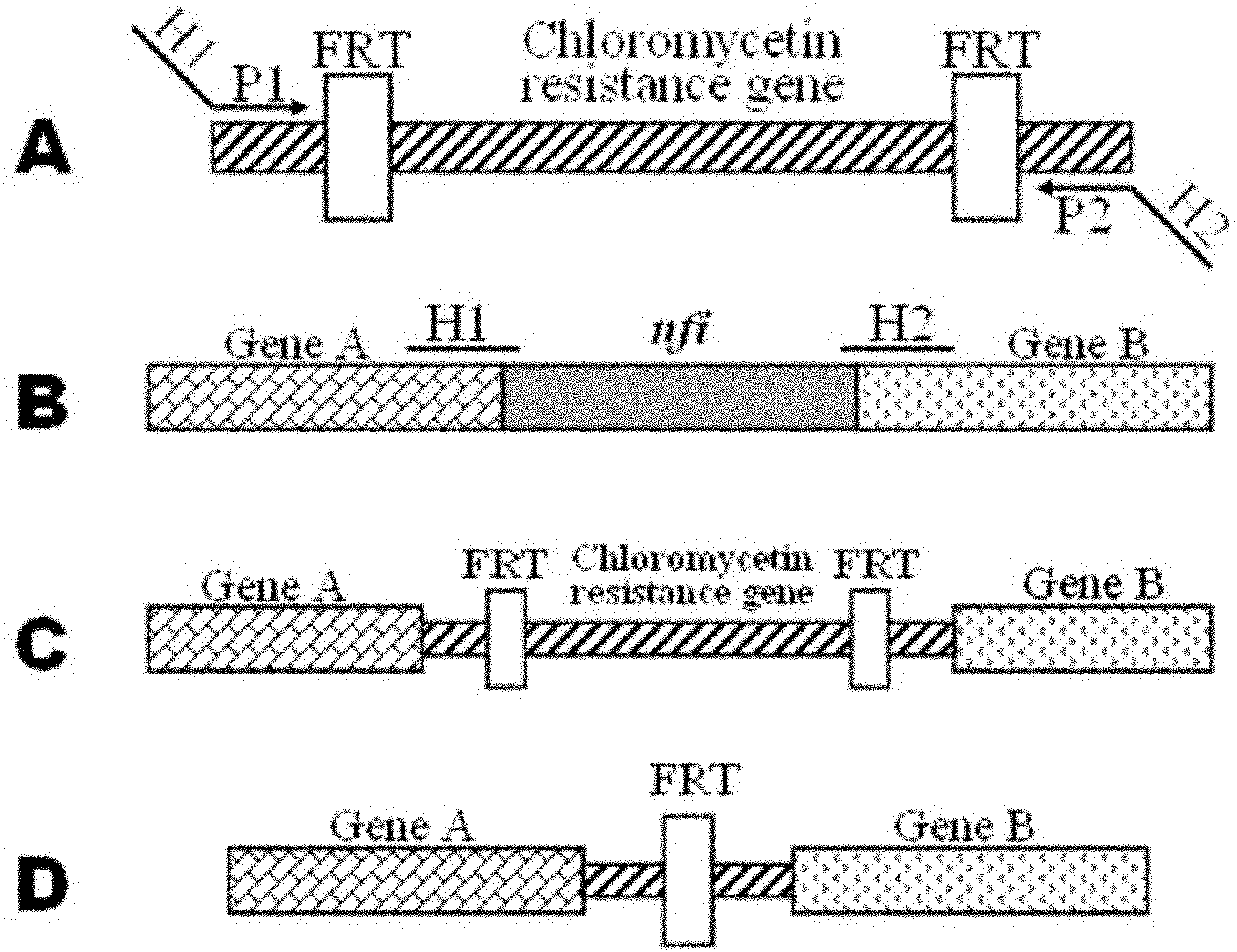
[0038] Example 1: Construction of linear gene targeting fragments
[0039] (1) Primer design: PCR primers were designed according to the DNA sequence of plasmid pKD3 (SEQ ID NO: 3 in the sequence table), and the base sequence is as follows:
[0040]Linear gene targeting primers (P7, P8):
[0041] P7: 5′ CGTGGAGGCAGTGCATCGACTGTCTGAACAGTATCACCGCTAAGGAGTGATTATG GTGTAGGCTGGAGCTGCTTC 3' (underlined is the 55bp upstream homology arm of the nfi gene, including only the start codon of the nfi gene)
[0042] P8: 5′ TTTGTAACATGTTGAGTTTCTCAAATACGGAAATTATCCGCAGTTTACCTGAATTA CATATGAATATCCTCCTTAG 3' (underlined is the 55bp downstream homology arm of the nfi gene, including only the stop codon of the nfi gene)
[0043] Primers P7 and P8 were used to construct linear gene targeting fragments.
[0044] (2) PCR amplification: using the DNA of plasmid pKD3 as a template, PCR was carried out with primers P7 and P8 to amplify the linear gene targeting fragment containing the chloramphenicol ...
Embodiment 2
[0056] Example 2: Construction, screening and identification of Escherichia coli DH5α strain nfi gene knockout mutant
[0057] (1) Preparation of Escherichia coli DH5α Competent Cells by Electroporation
[0058] The recipient bacteria (DH5α) were inoculated in LB medium and cultured with vigorous shaking on a shaker at 37°C until OD600 = 0.5 (about 3 hours); the bacterial solution was quickly placed on ice to cool for 10 minutes, and refrigerated and centrifuged at 3000g for 5 minutes at 4°C ;Discard the supernatant, add 1500μl ice-cold 10% glycerol, use a pipette gun to gently pump up and down to mix well, resuspend the cells, and centrifuge at 3000g at 4°C for 5 minutes; discard the supernatant, add 750μl ice-cold 10% glycerol , gently pump up and down with a pipette to resuspend the cells, and centrifuge at 3000g at 4°C for 5 minutes; add 20 μl of ice-cold 10% glycerol, and gently pump up and down with a pipette to resuspend the cells Suspended, used immediately or immedia...
Embodiment 3
[0088] Example 3: Analysis of Practical Sequencing Applications
[0089]In order to test the actual effect of the nfi gene knockout mutant strain of Escherichia coli DH5α on sequencing, we randomly selected a segment of the "shotgun non-clonable fragment" on the genome of Pseudomonas aeruginosa phage PaP1, used DH5αΔnfi as the cloning host bacteria, and used The shotgun strategy was used to clone and build a library, and finally successfully spliced this fragment that could not be cloned by the traditional shotgun method.
[0090]
[0091]
[0092]
[0093]
[0094]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com