Enhanced promoter and method for producing L-lysine usinge the same
A promoter, aspartic acid amino technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, using vectors to introduce foreign genetic material, etc., to achieve high promoter activity, improve aminotransferase activity, and increase production efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Identification of the transcription initiation site of the aspB gene
[0053] In this example, an unknown transcription start site of the aspB gene was identified.
[0054]In order to identify the transcription initiation site of the aspB gene, the 5'-RACE (rapid amplification of 5' cDNA ends) technique (Sambrook, J., Russell, D.W. Molecular Cloning: a laboratory manual) was used 3 rd ed. 8.54). 5'-RACE is a method of cloning an unknown 5' upstream region using known mRNA, and this method is performed using 5'-Full Race core set (Cat. No. 6122) obtained from Takara Corporation.
[0055] In order to carry out the 5'-RACE experiment, 5 kinds of primers (Table 1, SEQ ID NO: 5) were synthesized based on the base sequence of the aspB gene (NCBI Accession No. to 9).
[0056] Table 1
[0057] Primer
SEQ ID NO:
aspB(race) / RT
P-ccaagacctgctcc (5' phosphorylated)
5
aspB(race) / F1
Tactcgcggtaagccttcg
6...
Embodiment 2
[0061] Embodiment 2: the construction of the recombinant vector that is used for promoter improvement
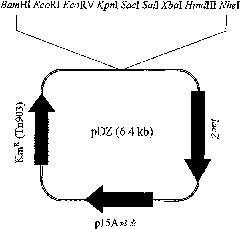
[0062] Construction of vector (pDZ) for chromosomal insertion
[0063] In this example, the vector pDZ for insertion into the chromosome of Corynebacterium was constructed based on the Escherichia coli cloning vector pACYC177 (New England Biolab, GenBank accession number #X06402).
[0064] The pACYC177 vector was treated with restriction enzymes Acul and BanI, followed by klenow treatment to obtain blunt ends. The lacZ gene of Escherichia coli origin used as a screening marker was amplified from the genomic DNA of Escherichia coli K12W3110 by PCR, the PCR was designed so that the amplified fragment included the promoter of the gene, and T4 DNA polymerase and polynucleoside Acid kinase treatment to generate blunt ends and phosphorylation of the 5' end. The above two DNA fragments are connected to each other to generate a circular DNA molecule, and then an artificially sy...
Embodiment 3
[0074] Embodiment 3: the recombinant vector is inserted in the Corynebacterium glutamicum bacterial strain
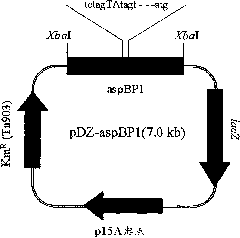
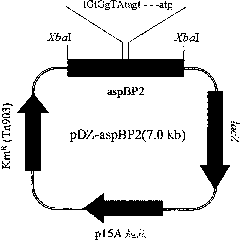
[0075] In this example, in order to improve the aspB promoter on the corynebacterium chromosome, the constructed recombinant vector was transformed into lysine-producing Corynebacterium glutamicum KFCC10881, and the promoter sequence of the vector was replaced by homologous recombination A promoter sequence on a chromosome such that an enhanced promoter sequence is inserted into the chromosome.
[0076] Each of the recombinant vectors pDZ-aspBP1, pDZ-aspBP2, and pDZ-aspBP3 for base substitution in Corynebacterium was designed to include a DNA fragment having the enhanced promoter sequence in Example 2, by electric pulse The above vector was transformed into Corynebacterium glutamicum KFCC10881 (transformed according to Appl. Microbiol. Biotechnol. (1999) 52: 541-545), and the transformant was screened on a medium containing 25 mg / L kanamycin to obtain Transformants i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com