Laodelphax striatellus lethal gene fragment Ribosomal protein L9-B based on gene-silencing technology and dsRNA thereof
A technology of gene fragments and striatellus, applied in the field of agricultural biology, can solve the problems of poor chemical control effect and environmental pollution, and achieve the effect of being convenient for a large number of experiments, convenient for experimental operations, and reducing mechanical damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] 1. Cloning method of Ribosomal protein L9-B gene fragment:
[0039] (1) Get 10-20 heads of SBPH, and use the TRIzol method to extract total RNA;
[0040] (2) Synthesizing the first strand of cDNA;
[0041] (3) Obtain the gene fragment sequence from the SBPH transcriptome, and after homology comparison at http: / / www.ncbi.nlm.nih.gov / , it is predicted to be the SBPH Ribosomal protein L9-B gene, Use Primer premier 5.0 software to design P 1 and P 2, and use RT-PCR method to amplify;
[0042] Upstream primer (P1): 5'TTCTAGCGAGTTTCCG 3' (SEQ ID NO.2),
[0043] Downstream primer (P2): 5'GCGATGCCCAGTATGT 3' (SEQ ID NO.3);
[0044] The PCR reaction program was: denaturation at 94°C for 2 min, 30 sec at 94°C, 30 sec at 55°C, 30 sec at 72°C, 35 cycles, and extension at 72°C.
[0045] PCR reaction system (50μL):
[0046]
[0047]
[0048] (4) The PCR product is separated by agarose gel electrophoresis, and the target DNA fragment is recovered;
[0049](5) Insert the re...
Embodiment 2
[0053] Embodiment 2.dsRNA synthesis and recovery
[0054] (1) According to the verified Ribosomal protein L9-B gene fragment sequence, Primer Premier 5.0 software was used to design P3 and P4, and the T7 promoter sequence TAATACGACTCACTATAGGG was added to the 5' end of the upstream and downstream primers;
[0055] Upstream primer (P 4): 5' TAATACGACTCACTATAGGG TTCTAGCGAGTTTCCG 3' (SEQ ID NO.5)
[0056] Upstream primer (P 5): 5' TAATACGACTCACTATAGGG GCGATGCCCAGTATGT 3' (SEQ ID NO. 6)
[0057]
[0058] PCR reaction program: denaturation at 94°C for 2min, 30sec at 94°C, 30sec at 60°C, 30sec at 72°C, 38 cycles, extension at 72°C.
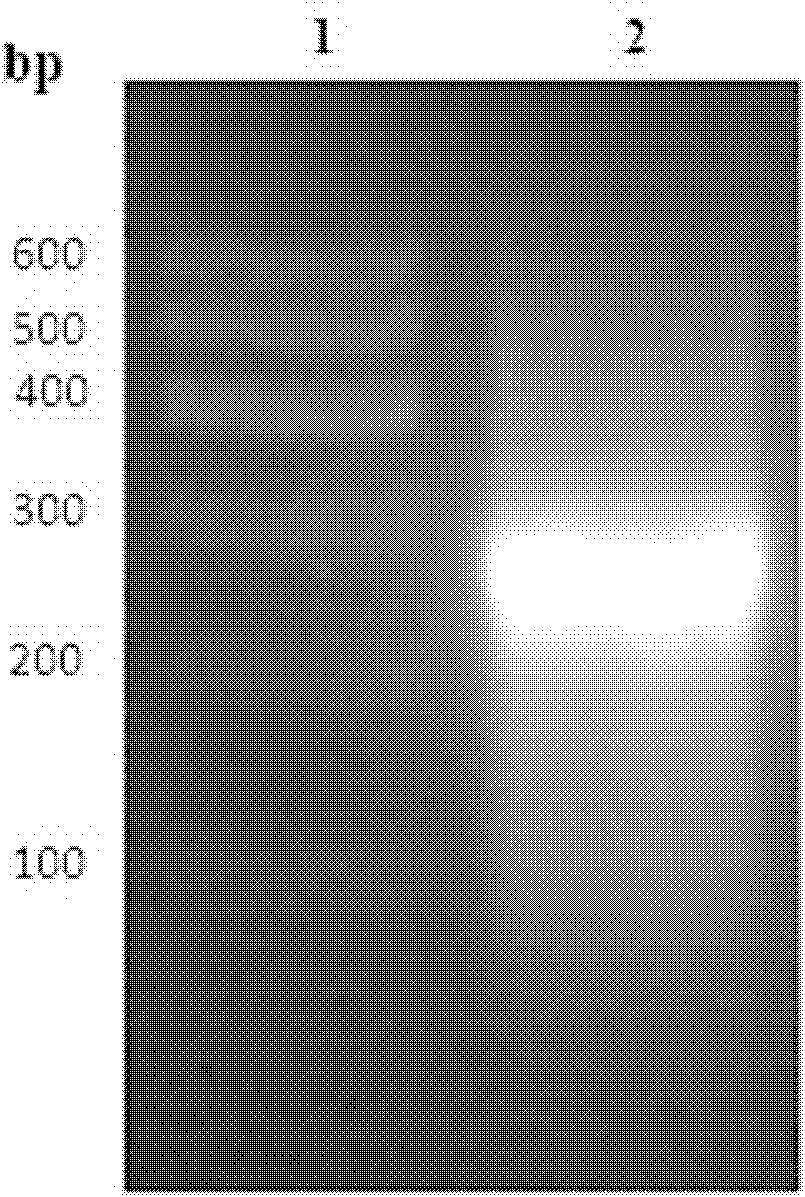
[0059] (2) The PCR product was separated by electrophoresis on a low-melting point agarose gel with a concentration of 1% and observed under ultraviolet light. The results are shown in figure 1 , whose sequence is shown in SEQ ID NO.4.
[0060] (3) Using Promega's SV Gel and PCR Clean-Up System kit for recovery:
[0061] ① Cut the gel of th...
Embodiment 3
[0071] Embodiment 3.dsRNA feeding experiment
[0072] (1) Seal one end of the glass tube with a parafilm, suck the second-instar SBPH into the glass tube with a sucker, and seal the other end with gauze;
[0073] (2) Gently pat the insects to one end with your hands, remove the gauze from the other end, put the prepared parafilm sticker up, pull evenly to both sides, pull it into a square, and then cover it on the glass On the nozzle of the tube, place the tube upright on the ultra-clean table;
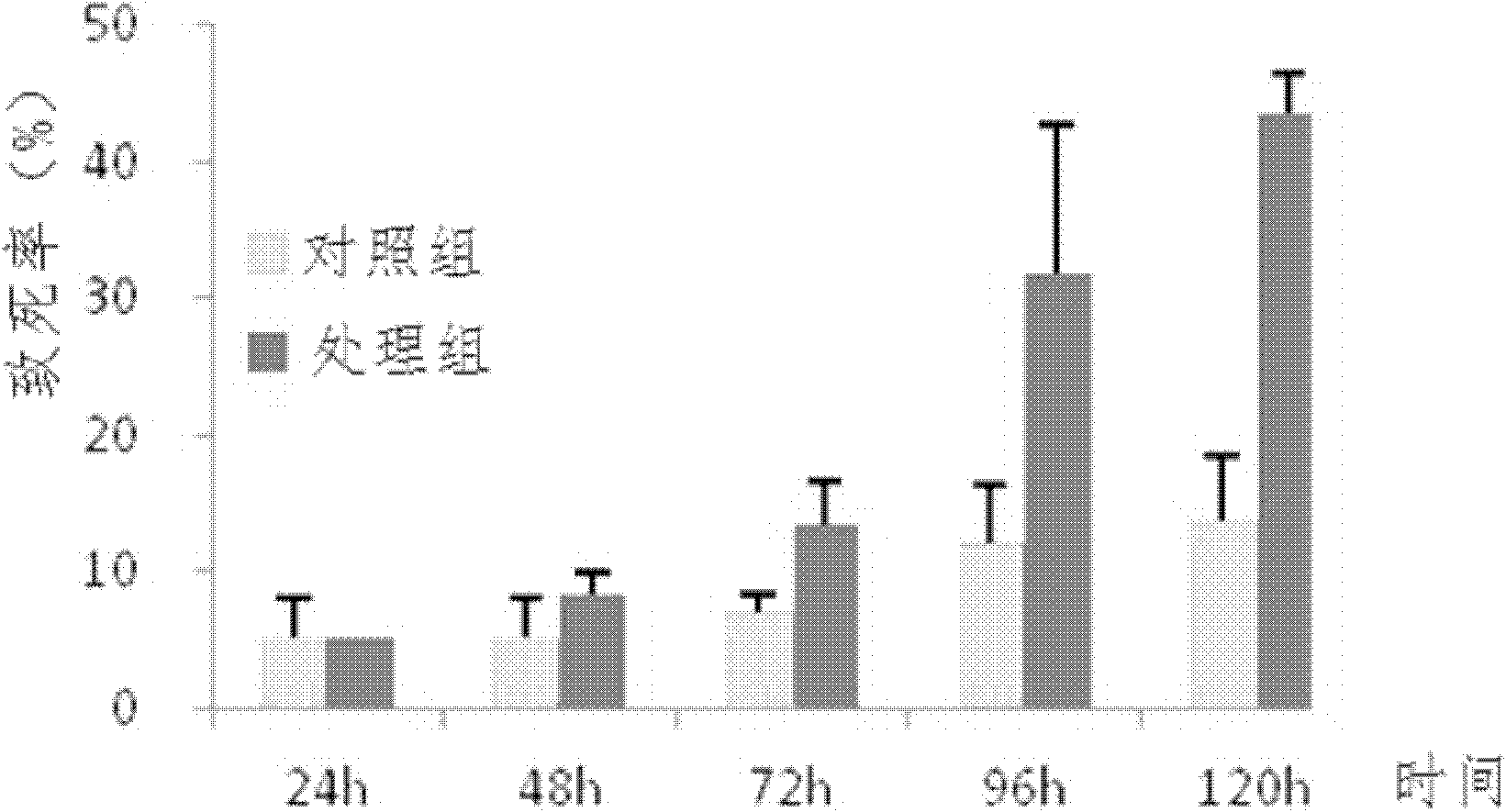
[0074] (3) Use a pipette gun to draw 100 μl of feed and drop it in the center of the parafilm. The control group only adds feed (see Table 1 for formula), and the treatment group adds dsRNA of Ribosomal protein L9-B gene in the feed, and the concentration of dsRNA is 3875ng / μl , with a new parafilm, with the sticker side facing down, stick it on the glass tube nozzle, and seal the feed and dsRNA between the two layers of parafilm;
[0075] (4) Put the glass tube with feed and dsRNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com