Method and kit for directly amplifying DNA (deoxyribonucleic acid) segment from biological material
A kit and direct technology, applied in the fields of molecular biology and genetic engineering, can solve problems such as difficulty in obtaining and loss, and achieve the effects of convenient operation, safe use, and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] (1) the solution adopted in embodiment 1
[0084] A. Sample lysate a, the sample lysate formula is
[0085] Lysis solution a-1: 70% PEG200; 20mM KOH, pH 13.5;
[0086] Lysis solution a-2: 60% PEG200; 20mM KOH, pH 13.5;
[0087] Lysis solution a-3: 60% PEG200; 40mM KOH, pH 13.8;
[0088] Lysis solution a-4: 50% PEG200; 20mM KOH, pH 13.5.
[0089] B. Neutralizing solution, the formula is as follows
[0090] 200mM Tris-HCl (pH 6.8-7.5).
[0091] C.2×Taq PCR mixture, the formula is as follows
[0092] 2×PCR Buffer;
[0093] 3mM MgSO 4 ;
[0094] 0.2mM dNTPs;
[0095] 0.1U / μL Taq DNA polymerase;
[0096] 2×Loading Buffer.
[0097] D.2×PCR Buffer, the formula is as follows:
[0098] 40mM Tris-HCl, pH=8.825°C;
[0099] 20mM (NH4) 2 SO 4 ;
[0100] 20mM KCl;
[0101] 0.2%Trtion X-100;
[0102] 0.2mg / mLBSA.
[0103] E.2×Loading Buffer, the formula is as follows:
[0104] 8% Glycerol;
[0105] 0.05% Bromophenol Blue.
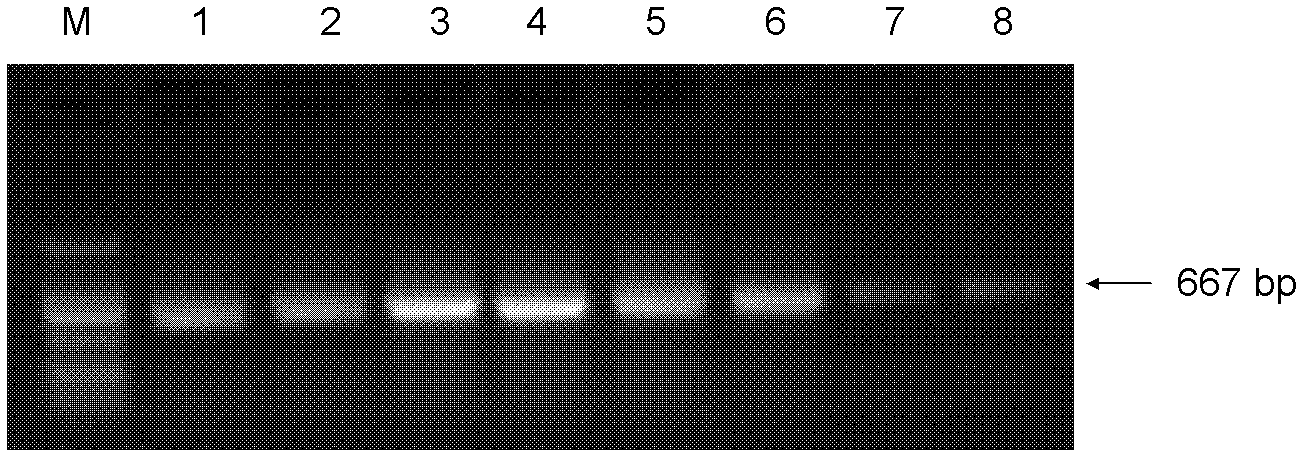
[0106] (2) Genomic DNA of tobacco was e...
Embodiment 2
[0119] (1) the solution adopted in embodiment 2
[0120] A. Sample lysate b, the sample lysate formula is
[0121] Lysis solution b-1: 5% chelex-100 (chelex-100 was purchased from sigma company)
[0122] Lysis solution b-2: 10% chelex-100 (chelex-100 was purchased from sigma company)
[0123] B. 2×Taq PCR mixture, the formula is as follows
[0124] 2×PCR Buffer;
[0125] 3mM MgSO 4 ;
[0126] 0.2mM dNTPs;
[0127] 0.1U / μL Taq DNA polymerase;
[0128] 2×Loading Buffer.
[0129] The 2×PCR Buffer and 6×Loading Buffer used therein are exactly the same as those in Example 1.
[0130] (2) Genomic DNA is extracted from a small amount of blood.
[0131] 1. Take 5-10 μL of anticoagulated whole blood and add it to a 1.5ml centrifuge tube, add 500 μl of sterile water, and shake vigorously.
[0132] Centrifuge at 12,000rpm for 2min, discard the supernatant, and collect the precipitate.
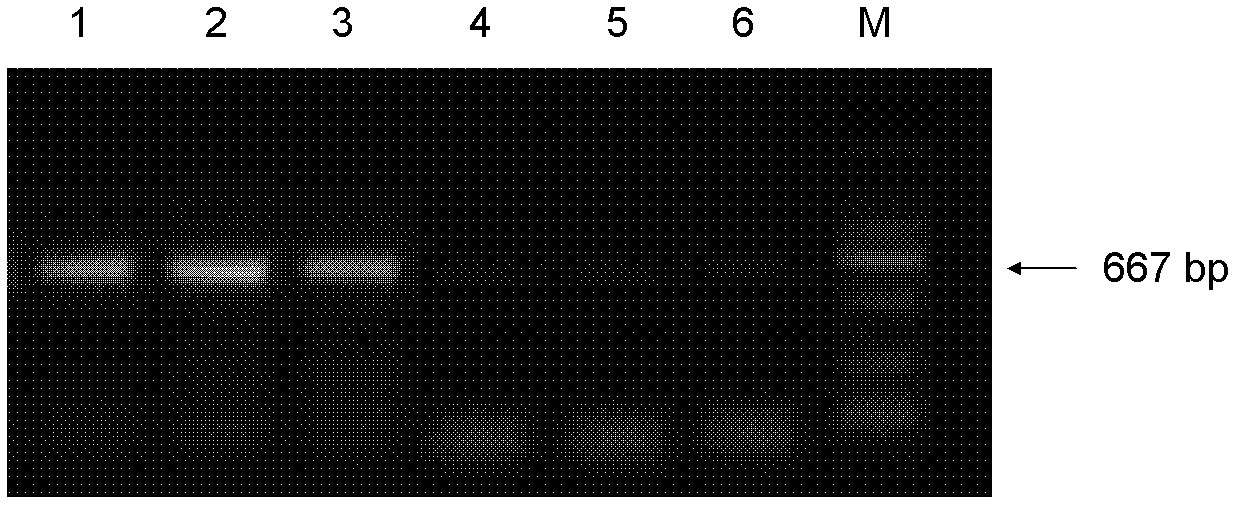
[0133] 2. Add 100 μL Lysis Solution 2 to the precipitation, bathe in water at 56°C for 30 m...
Embodiment 3
[0143] (1) the solution adopted in embodiment 3
[0144] A. Sample lysate c, the sample lysate formula is
[0145] Lysis solution c-1: 10% chelex-100, 1% TritonX-100;
[0146] Lysis buffer c-2: 10% chelex-100, 2% TritonX-100.
[0147] B. 2×Taq PCR mixture, the formula is as follows
[0148] 2×PCR Buffer;
[0149] 3mM MgSO 4 ;
[0150] 0.2mM dNTPs;
[0151] 0.1U / μL Taq DNA polymerase;
[0152] 2×Loading Buffer.
[0153] The 2×PCR Buffer and 6×Loading Buffer used therein are exactly the same as those in Example 1.
[0154] (2) Genomic DNA extraction from hair.
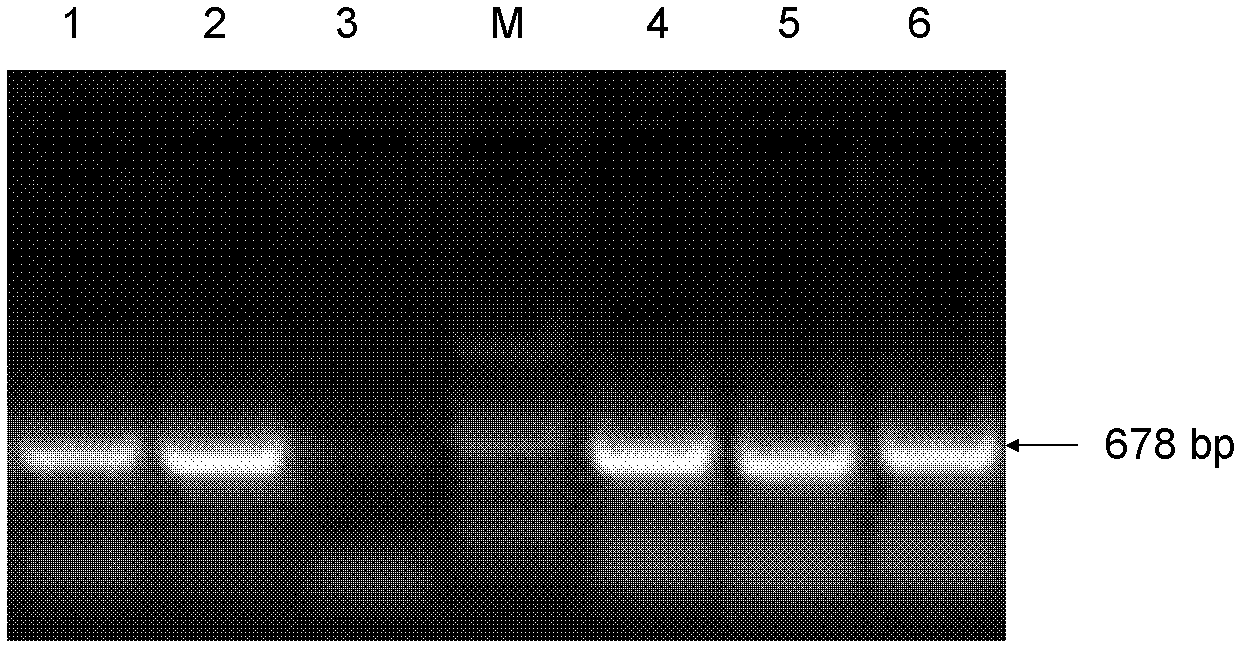
[0155] 1. Take 3-4 hairs with hair follicles, take the roots, put them into a 1.5ml centrifuge tube, add 30μl lysate c and 1μL Proteinase K, shake, 56°C water bath for 30min, 100°C water bath for 8min.
[0156] 2. Centrifuge at 12,000 rpm for 3 minutes, and take the supernatant for PCR reaction.
[0157] (3) PCR reaction
[0158] The template obtained above was subjected to PCR amplification. The upstream and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com