Specific mark of thinopyrum elongatum chromosome in wheat background and application thereof
A technology of E. elongatum and specific markers, applied in the field of crop genetics and breeding, to achieve superior stability, excellent marker resources, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1 plant material and PCR primer sequence
[0069] (1) Plant material
[0070] The materials used in the present invention include 7 parts of Chinese spring wheat-Thinopyrum elongatum disomic additional lines (Triticum aestivum cv. Chinese Spring-Thinopyrum elongatum disomic additional lines, DA1E, DA2E, DA3E, DA4E, DA5E, DA6E, DA7E,), 2 parts Chinese spring wheat-Thinopyrum elongatum 7E long arm and 7E short arm addition lines (DA7ES, DA7EL), 19 Chinese spring wheat-Thinopyrum elongatum disomic substitution lines (Triticum aestivum cv.Chinese Spring-Thinopyrum elongatum disomic substitutional lines, Such as DS1E / 1A, DS2E / 2A, DS3E / 3A to DS3E / 19A, 1 part of wheat Chinese spring (Triticum aestivum) and 1 part of diploid Elongated wheatgrass. The above materials are donated and preserved by Dr.Fedax of the Canadian Department of Agriculture In the Laboratory of Molecular Cytogenetics, School of Biotechnology, Yangzhou University.
[0071] (2) PCR amplification ...
Embodiment 2
[0077] The extraction of embodiment 2 genome DNA
[0078] DNA was micro-extracted by the SDS phenol-chloroform method. The steps are as follows:
[0079] (1) Take the young leaves (about 0.1g), cut them into pieces and put them into a 2ml centrifuge tube, cool them in liquid nitrogen, and grind them to powder with a grinding rod;
[0080] (2) Place the centrifuge tube at room temperature to cool slightly, add 700 μl of buffer A, mix gently, then bathe in water at 65°C for 20 minutes, and mix by inverting up and down every 5 minutes;
[0081] Buffer A:
[0082]
[0083] Dilute the volume to 1L with ddH2O and use it after sterilization.
[0084] (3) Take it out and cool it down to room temperature, add 350 μl of phenol and chloroform each, turn it upside down, mix well, and extract for 5 minutes;
[0085] (4) 12000rpm, centrifuge for 10min, draw the supernatant into a new centrifuge tube;
[0086] (5) Add about 750 μl of chloroform, turn it upside down, mix well, and ext...
Embodiment 3
[0092] Example 3 PCR Amplification of Specific Fragment of Echinopsis elongatum
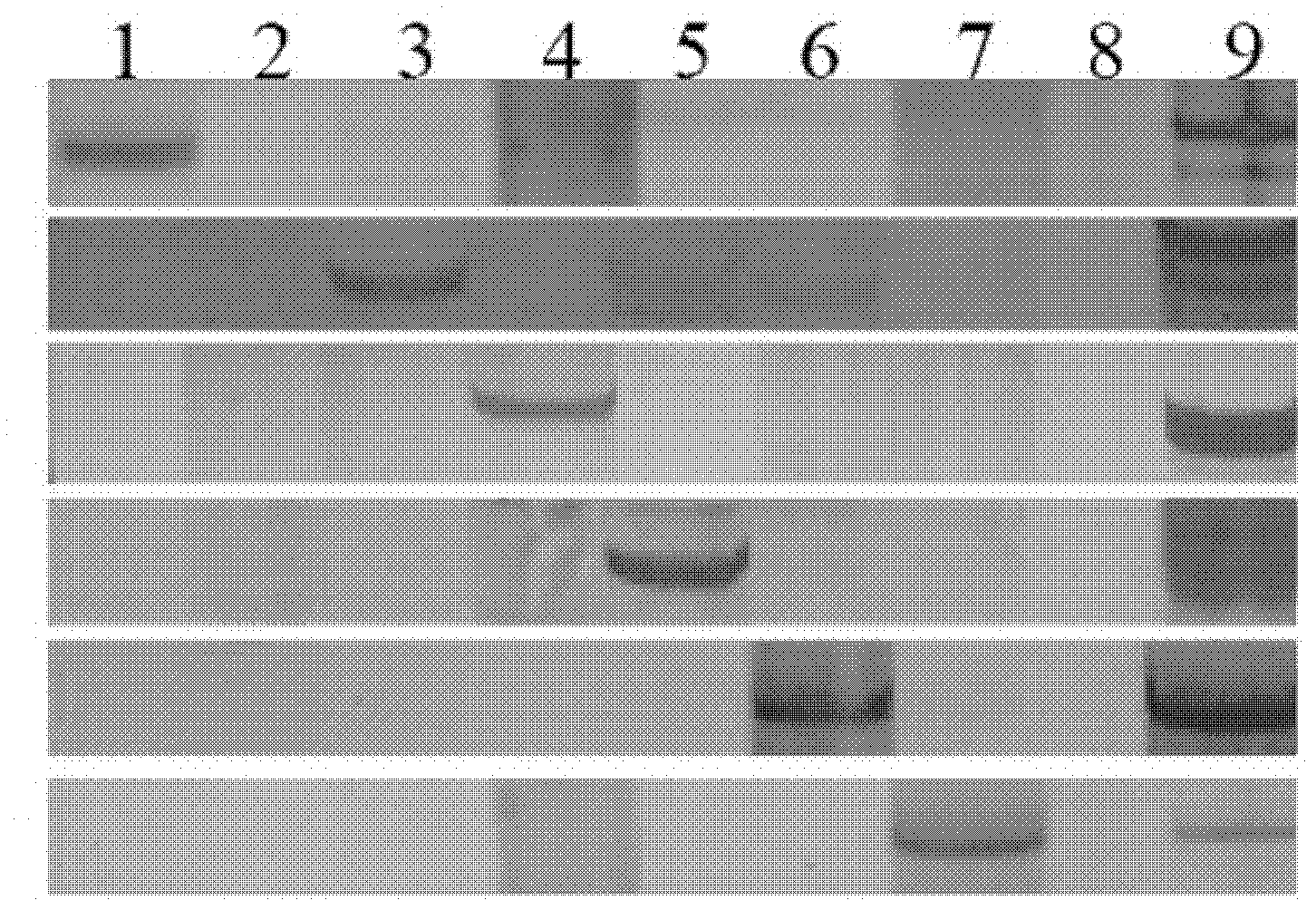
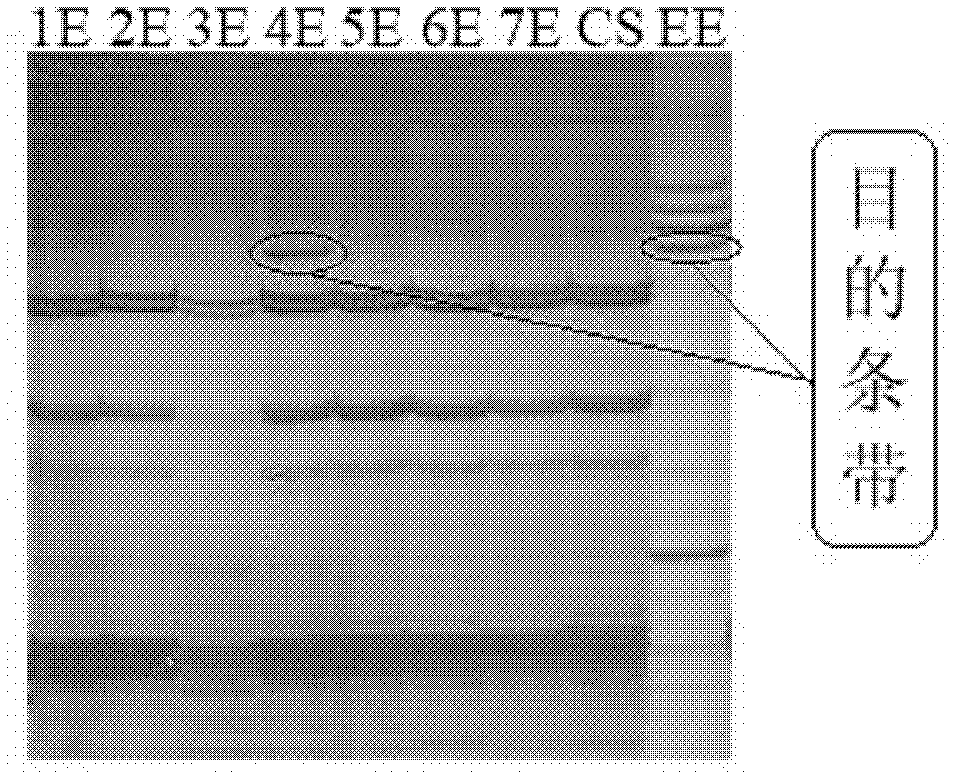
[0093] According to the combination of basic primers in Table 1, PCR amplification was carried out. The specific reaction system and reaction program of PCR are shown in Table 3 and Table 4. After the PCR reaction, the PCR products were detected by 6% polyacrylamide gel electrophoresis ( figure 1 ), the specific fragment sizes and specific chromosomes of E. elongatum amplified by each primer combination are shown in Table 2. Since there is a complete set of Chinese spring-Thinopyrum elongata disomic addition line materials, the amplified specific fragments can be further mapped to different E chromosomes.
[0094] Table 3 PCR reaction system
[0095]
[0096] Table 4PCR reaction program
[0097]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com