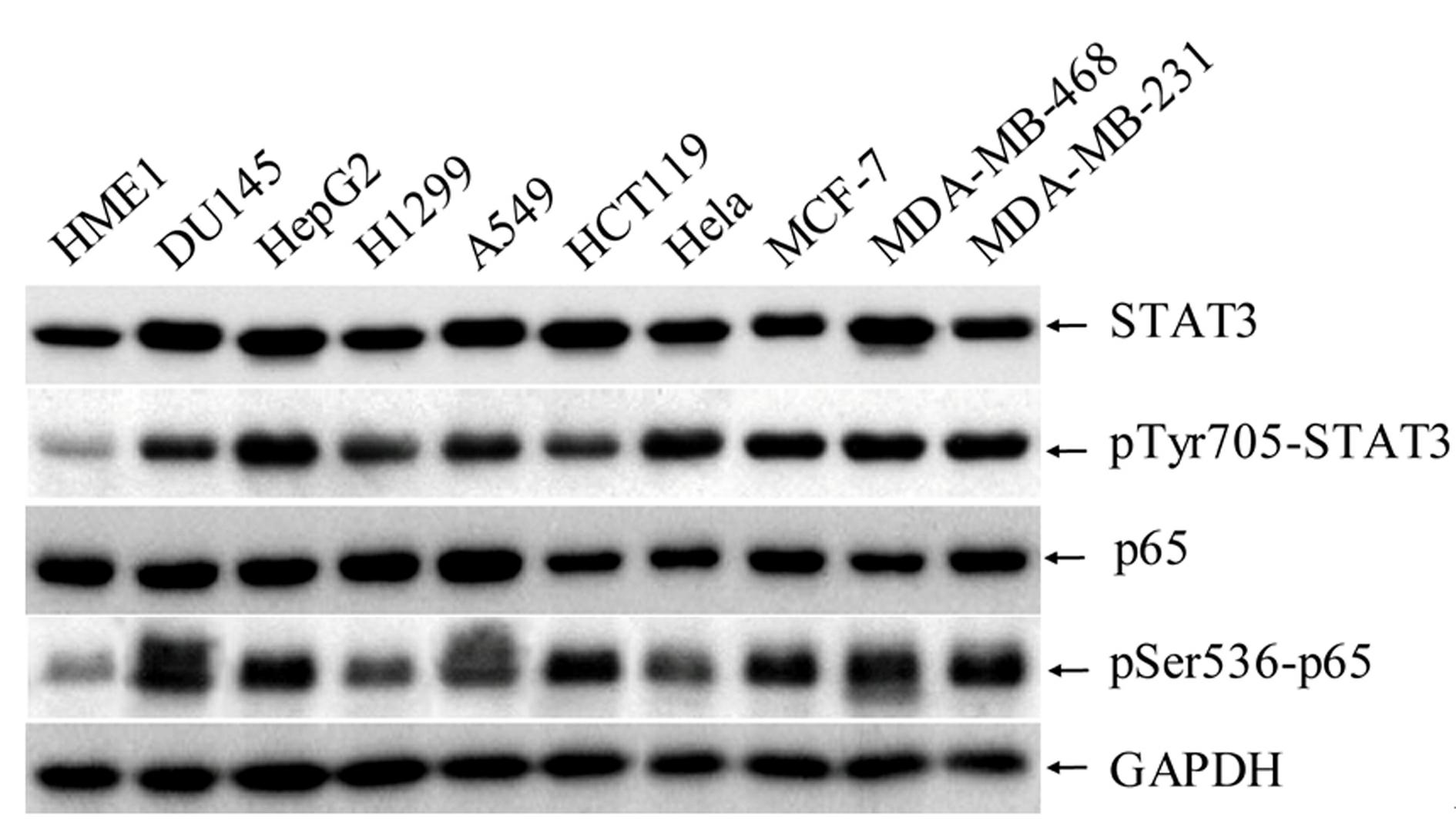
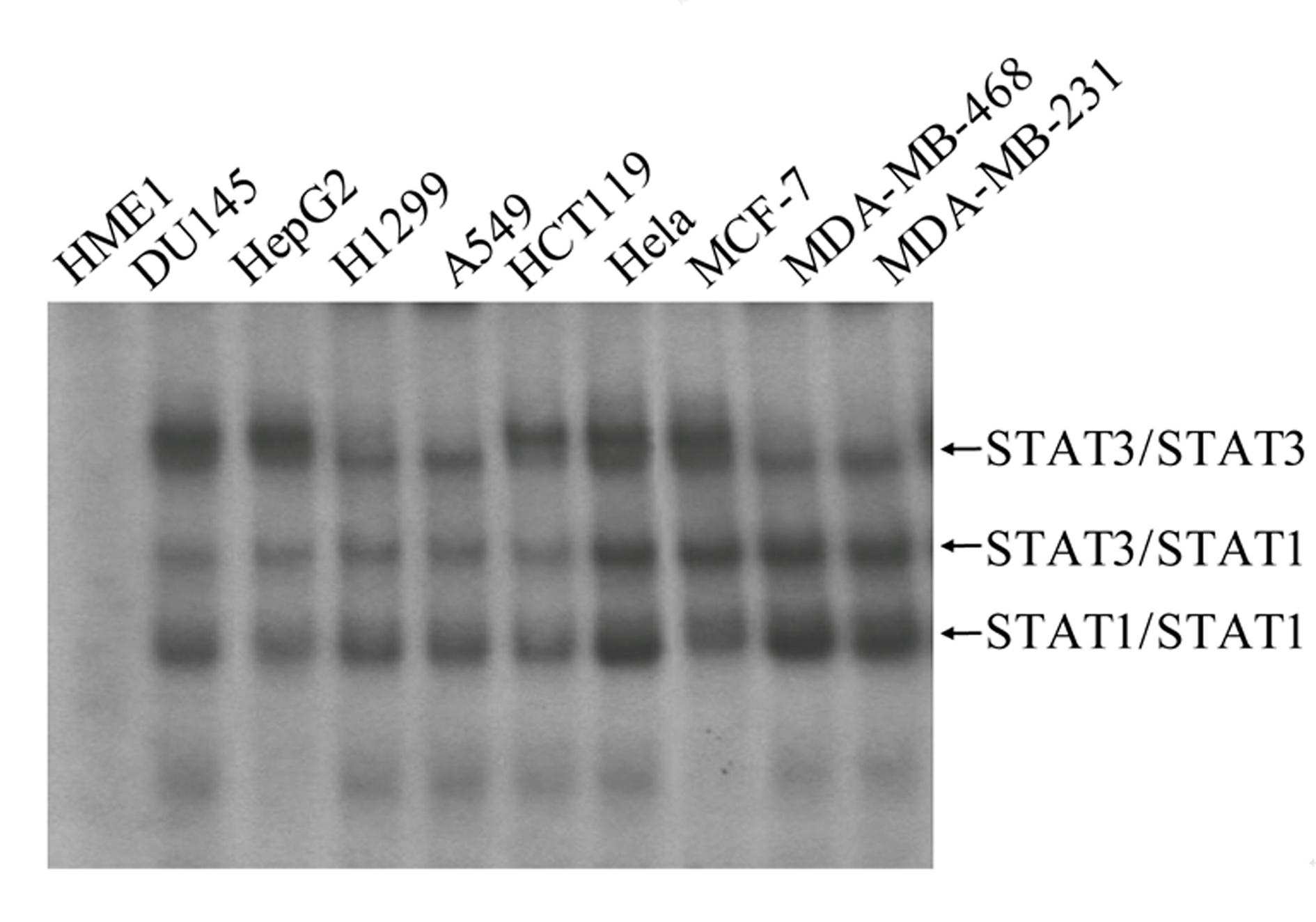
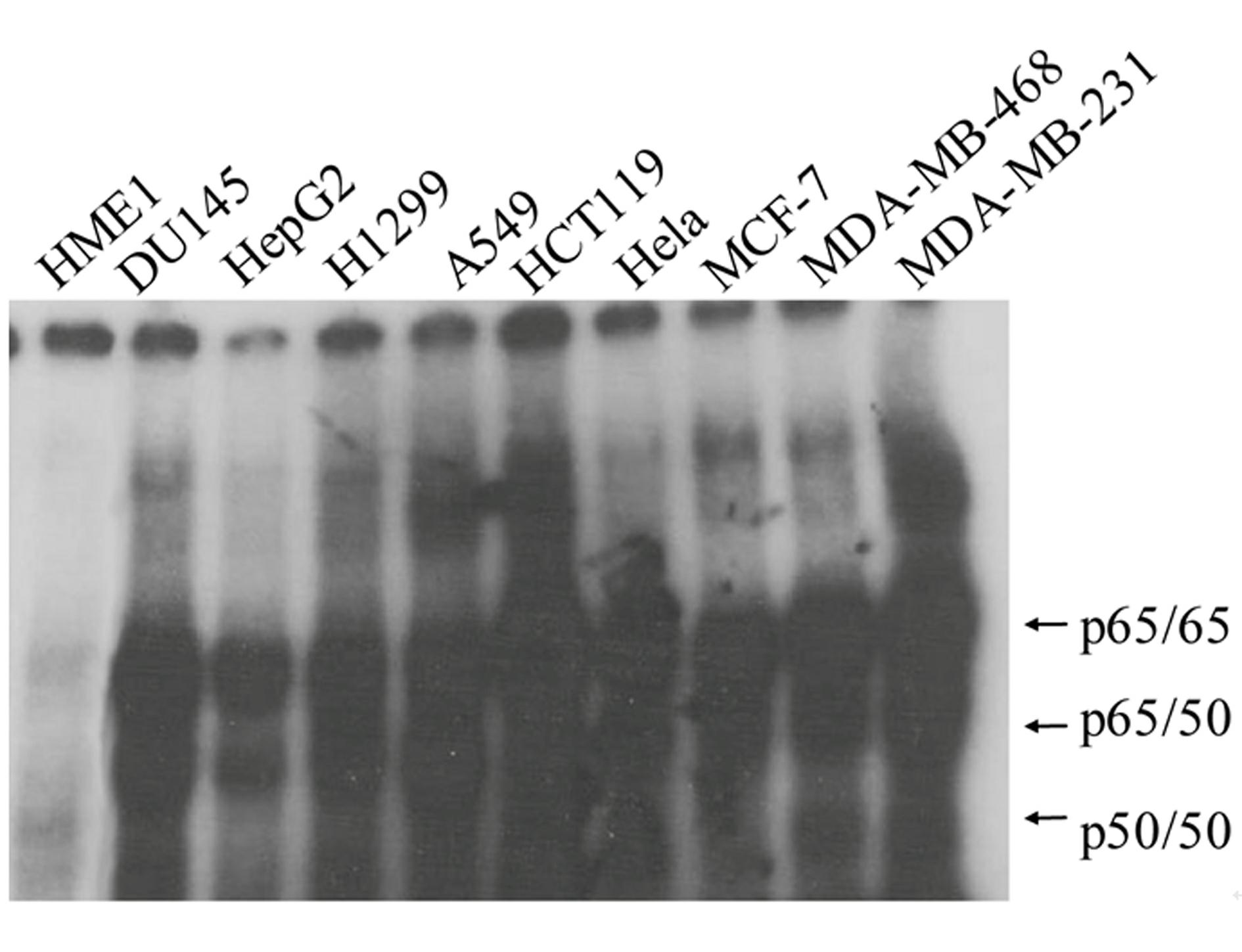
High-flux anti-tumor drug screening cell model based on STAT3 and NF-kB two-signal channel serving as target, as well as building and application of high-flux anti-tumor drug screening cell model
A technology of anti-tumor drugs and cell models, which is applied in the construction of anti-tumor drug screening cell models and high-throughput anti-tumor drug screening cell models. It can solve the problems of incomplete simulation of tumor cells, increased system instability, and increased costs. problem, to achieve the effect of shortening the screening cycle, reducing operation steps and improving stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] 1. Construction of reporting system carrier
[0050] according to Figure 4 As shown in the schematic diagram, the response sequence is artificially constructed, which includes STAT3 binding sequence and NF-κB binding sequence. The STAT3 binding sequence is referred to as SIE, and the NF-κB binding sequence is referred to as κB. The STAT3 binding sequence here consists of 5'-TTCCCGTAA-3' and 5'-TTCCTGTAA-3' 8 alternating series; the NF-κB binding sequence is 5'-GGGAATTTCC-3'; the two binding sequences are connected in series, Then add the general TATA box auxiliary sequence 5'-TATATAA-3', insert it into the general luciferase reporter vector pBR322-luc-puro, and the recombinant vector is named SIE-κB-luc; as a control, STAT3 will be included alone Nucleic acid sequences of the binding sequence or NF-κB binding sequence (the rest are the same) are inserted into the luciferase reporter vector, named SIE-luc and κB-luc, respectively. The ratio of the two response elemen...
Embodiment 2
[0065] 1. Construction of reporting system carrier
[0066]①Use the SIE-κB-luc vector in Example 1 to extract with the Tiangen High Purity Plasmid Mini-Extraction Kit, take 10 μg for double enzyme digestion with XhoⅠ and BglⅡ (3 μl each), 37 °C, 8 h, Agarose gel electrophoresis, using the Tiangen DNA Purification and Recovery Kit to recover the product I, the product at this time excised the κB sequence; two DNAs with cohesive ends (and can be paired with XhoI and BglII restriction sites) Complementary primers, including two SIE sequences 5'-TTCCCGTAA-3' and two κB sequences 5'-GGGAATTTCC-3', see underlined)
[0067] 5'-TCGAGC GGAAATTCCCGTAA AC GGAAATTCCCGTAA AA-3', and
[0068] 5'-GATCTT TTACGGGAATTTCC GT TTACGGGAATTTCC GC-3'
[0069] Dilute to a 50 μM solution, mix 15 μl each, put in a boiling water bath at 95 °C for 5 minutes, and cool naturally in the water bath to obtain product II; Ligation, ligation overnight at 16°C; ③Transform Escherichia coli competent DH5...
Embodiment 3
[0081] 1. Construction of reporting system carrier
[0082] Using the same primer annealing and enzyme-cut ligation methods as in Example 1, the complete insert sequence inserted into the multiple cloning site of the luciferase reporter vector pBR322-luc-puro is as follows:
[0083]
[0084] 2. Report system carrier performance test
[0085] Same as Example 2, but when 293T cells are used to detect the simultaneous activation of STAT3 and NF-kB signaling pathways, the fluorescence value that can be achieved by the reporter system is only about 37% greater than the sum of activation of individual signaling pathways, and the fluorescence value increased by STAT3 activation is only Adding about 10% of the fluorescence value for NF-kB activation, the decrease in fluorescence value when using the same inhibitor to inhibit the STAT3 signaling pathway is not as obvious as the reporter system vectors in Examples 1 and 2.
[0086] 3. Drug screening model cell selection
[0087] Wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com