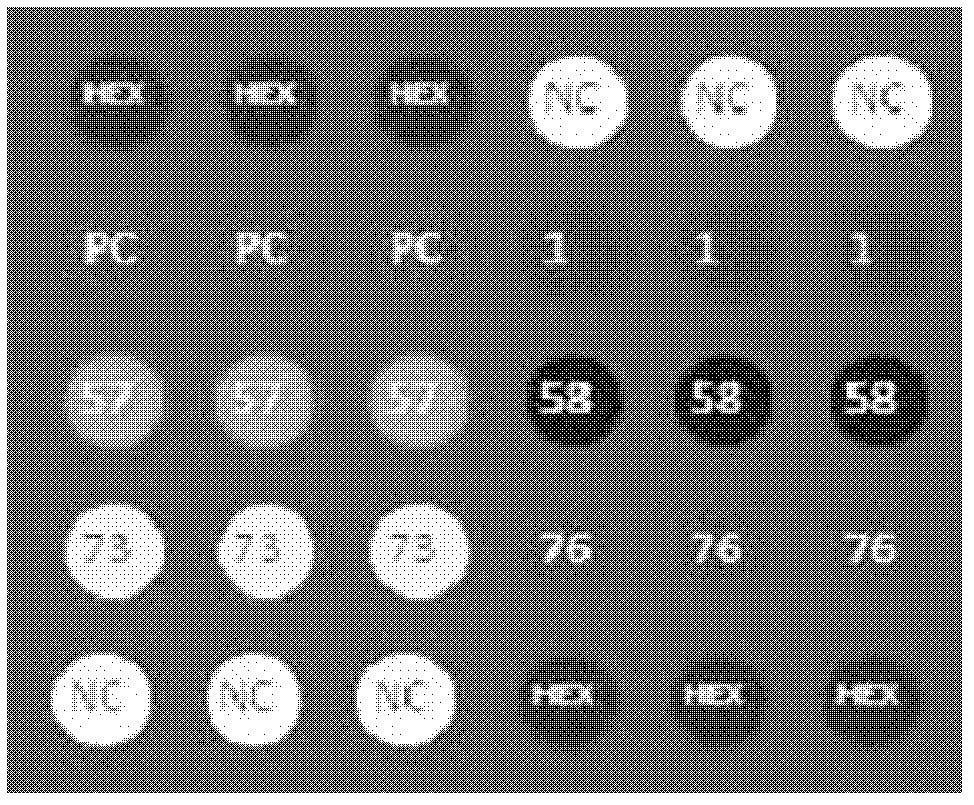
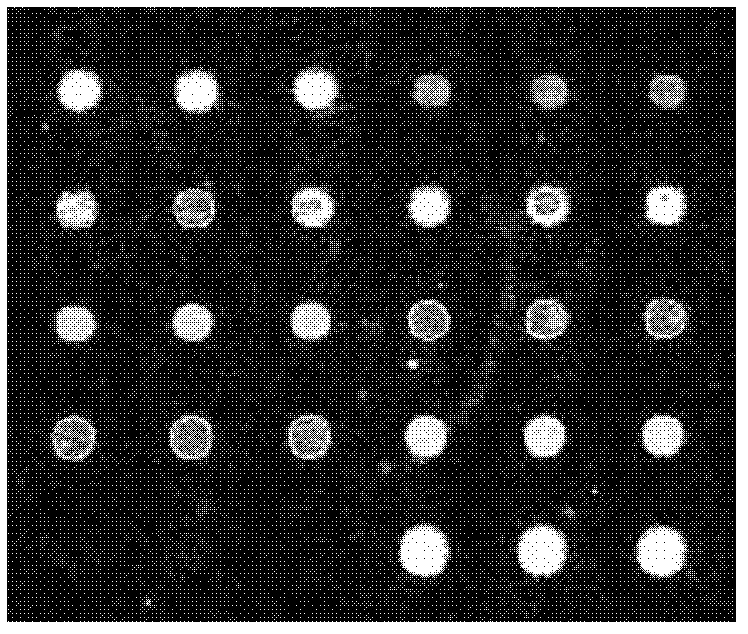
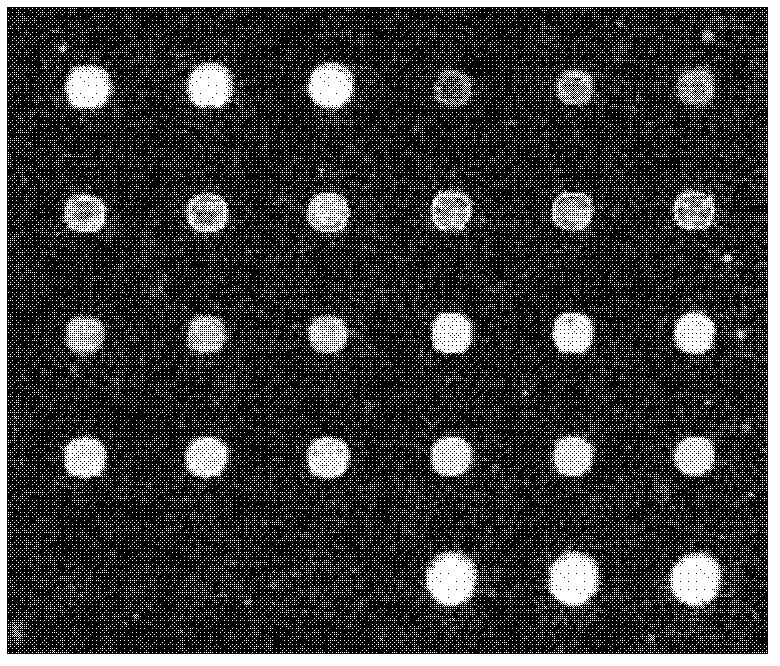
Chip for screening 16SrI group phytoplasmas and application thereof
A phytoplasma and gene chip technology, applied in the fields of bioinformatics and bio-quarantine identification, can solve the problems of low specificity, inability to detect and monitor phytoplasma, cumbersome steps, etc., and achieve good results and intuitive analysis results Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Embodiment 1, the preparation of the chip of screening 16SrI group phytoplasma
[0072] 1. Design of group-level highly compatible oligonucleotide probes
[0073] 1) Download the 16SrI group phytoplasma genome and nucleic acid sequence data from the National Center for Biotechnology Information (NCBI) and the official website of the Ribosome Database Project (RDP) database for designing probes.
[0074] 2) Organize phytoplasma nucleic acid sequences, remove nucleic acid sequences less than 200 bp in length and group them; all phytoplasma sequences in the same group are referred to as in-group sequences, and all phytoplasma sequences not in this group are referred to as out-group sequences.
[0075] 3) Use ClusterW to align the sequences in the group, find the sequence conserved regions and design probes. When designing the probe, use 2 to 3 bases as the interval in the conserved region of the sequence, continuously extract all 19 bp nucleic acid sequences, and screen t...
Embodiment 2
[0086] Embodiment 2, the application of the chip of screening 16SrI group phytoplasma
[0087] 1. Chip detection samples for screening 16SrI group phytoplasma
[0088] 1. Extraction of total DNA from samples used for testing
[0089] 1) get the strawberry leaf (Latin name: Strawberry Phylloid Fruit Phytoplasma, recorded in: Molecular Identification and Classification of Strawberry Phylloid Fruit Phytoplasma in Group 16SrI, New Subgroup.Plant disease, 2002, 86 (8) that infects strawberry branch fruit phytoplasma : 920., the public can obtain from the Chinese Academy of Inspection and Quarantine.) and Paulownia leaves infected with Paulownia witches'broom phytoplasma. Preparation and application, Acta Phytopathology, 2011, 41(2): 161-170.) each 0.1g, take an appropriate amount of liquid nitrogen to grind, then add 2ml of grinding liquid to fully grind, centrifuge at 20000rpm at 4°C for 20min, discard the supernatant.
[0090] 2) Add 1ml DNA extraction solution, 40ul proteinase...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com