Plasma miRNA (micro Ribonucleic Acid) profile and assay kit for predicting curative effect of interferon on treatment of chronic hepatitis B
A technology for chronic hepatitis B and interferon, which is applied in the fields of molecular biology and medicine, can solve problems such as limited prediction ability, and achieve the effects of low cost, convenient detection method, and reduced treatment cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Detection of miRNA expression profiles
[0026] 1. Experimental materials:
[0027] The invention obtains plasma and serum samples from 94 cases of chronic hepatitis B patients receiving long-acting interferon and common interferon. This study was approved by the Ethics Committee, and all patients signed informed consent. The training set included 66 HBV patients who received EG-IFN α2a (180μg / week) or PEG-IFN α2b (100μg / week), and the test set included 28 patients who received common interferon (IFN-α2b or IFN-α1b, 3-5MU / qod) HBV patients. The basic clinical data of the enrolled patients are shown in Table 1, 81.9% of them were positive for e antigen, and the average viral load was 7.05 log 10 copies / ml. Inclusion and exclusion criteria: The enrolled HBV patients had not received nucleoside analogs or interferon therapy 6 months before starting treatment, and the viral load was 5×10 4 copies / ml and above, abnormal liver alanine aminotransferase (>1...
Embodiment 2
[0043] Embodiment 2: Statistical analysis and establishment of forecasting model
[0044] 1. Test method:
[0045] Sorting of eigenvalues: the present invention uses the OneR method in the weka software package to sort miRNAs. The OneR method scores each feature by comparing the frequency table of feature values and the correlation of treatment effects.
[0046] Support Vector Machine (SVM)
[0047] Support Vector Machine (SVM) is evolved from statistical learning algorithm and is one of the most effective methods among many complex binary classification algorithms. In this study, R software was used to train and test the SVM classification model.
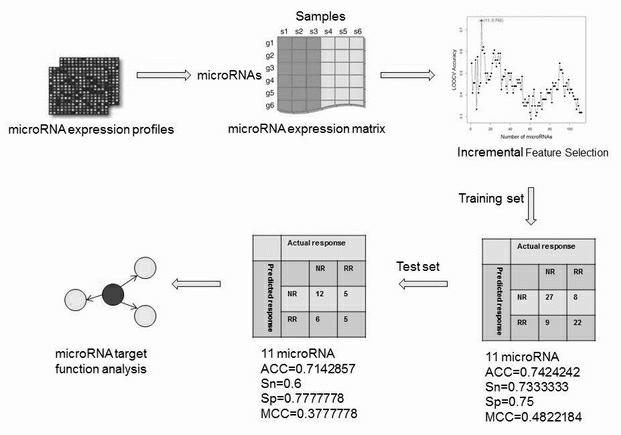
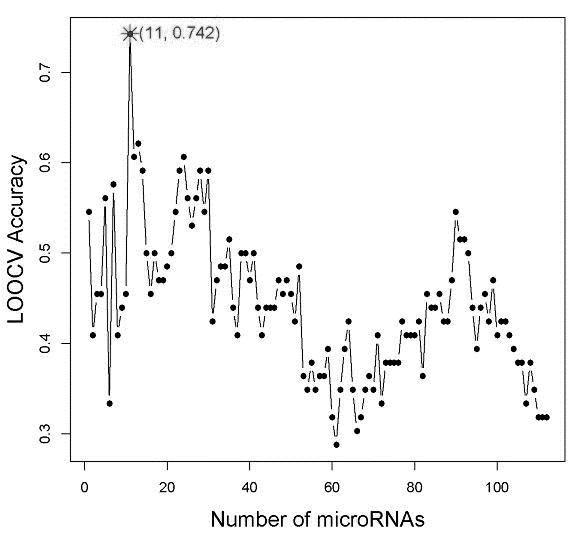
[0048] The present invention uses a leave-one-out cross-validation method to evaluate the performance of the prediction model. In this method, each data is sequentially removed as test data for other data, and the prediction engine is trained through multiple detections. The final total forecast accuracy is calculated as f...
Embodiment 3
[0062] Example 3 Prediction of 28 samples receiving common interferon therapy
[0063] The present invention further tested whether the prediction model of these 11 miRNAs could effectively distinguish the initial virological response in a test set consisting of 28 patients. As shown in Table 1, the basic clinical data of the test set and the training set are similar, except that the patients in the test set received common interferon treatment. Since different interferon preparations will cause differences in the effective drug concentration in vivo, this may affect the performance of the prediction. Nevertheless, we still get an overall prediction accuracy of 71.4% on the training set.
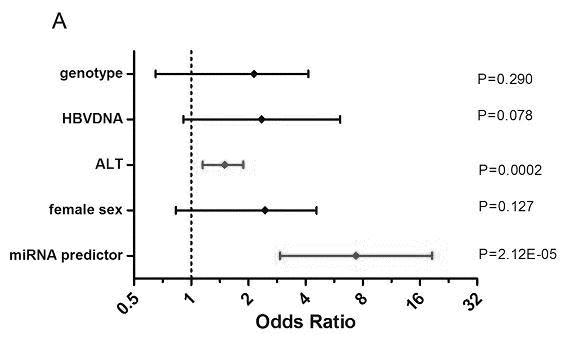
[0064] Comparison of miRNA prediction models with known factors associated with efficacy
[0065] The present invention further carried out univariate and multivariate Logistic regression analysis to evaluate whether the miRNA model is an independent predictor related to curative effect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com