Method for designing siRNA (Small Interfering Ribonucleic Acid) molecule resisting RNA virus
A technology of RNA virus and design method, applied in the field of RNA interference, can solve the problems of inefficiency, achieve a high degree of conservation, reduce the cost of anti-virus research and application, and facilitate the promotion and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Design and synthesis of small RNA molecules
[0022] (1) select the coding genes of TGEV structural protein and RdRP respectively;
[0023] (2) Using software to generate anti-TGEV siRNA molecules.
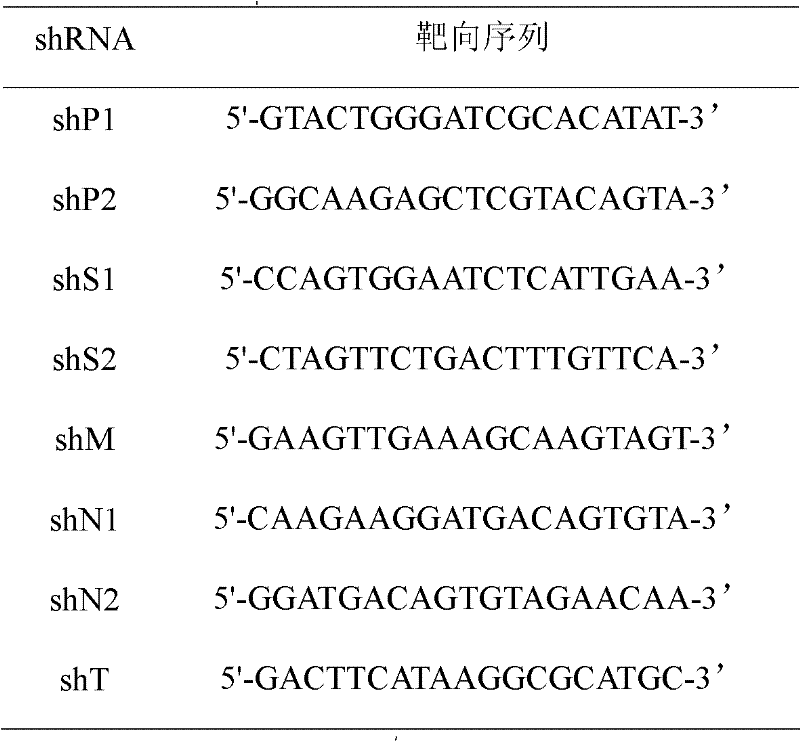
[0024] (3) According to the published small RNA sequence design rules, theoretically efficient siRNA molecules were selected and constructed into hairpin-type small RNA (shRNA) molecules (Table 1).
[0025] (4) The shRNA molecule was inserted into the vector to construct a plasmid expression vector containing U6 promoter.
Embodiment 2
[0027] Transfection of shRNA expression plasmids into ST cells
[0028] (1) ST cells (2-3×10 4 each / well) were inserted into a 48-well plate and cultured in complete DMEM;
[0029] (2) To the transfection reagent TransFast TM Add 400 μl DNase-free water to the transfection reagent, vortex to dissolve, and store at -20°C;
[0030] (3) 1.2 μg of dissolved plasmid DNA was dissolved in 300 μl of serum-free and double-antibody-free medium;
[0031] (4) Melt the transfection reagent at room temperature and vortex, add 3.6 μl to the DNA / DMEM mixture, vortex and mix; incubate at room temperature for 10-15 minutes to form a transfection complex;
[0032] (5) Remove the medium in the ST cells, add 100 μl / well of the transfection complex, and continue to store at 37°C in 5% CO 2 Incubate in the incubator for 1 hour;
[0033] Add 400 μl double antibody-free complete medium (containing 5% NBS) to ST cells; at 37°C, 5% CO 2 continue to grow in the incubator.
Embodiment 3
[0035] TGEV infection of ST cells
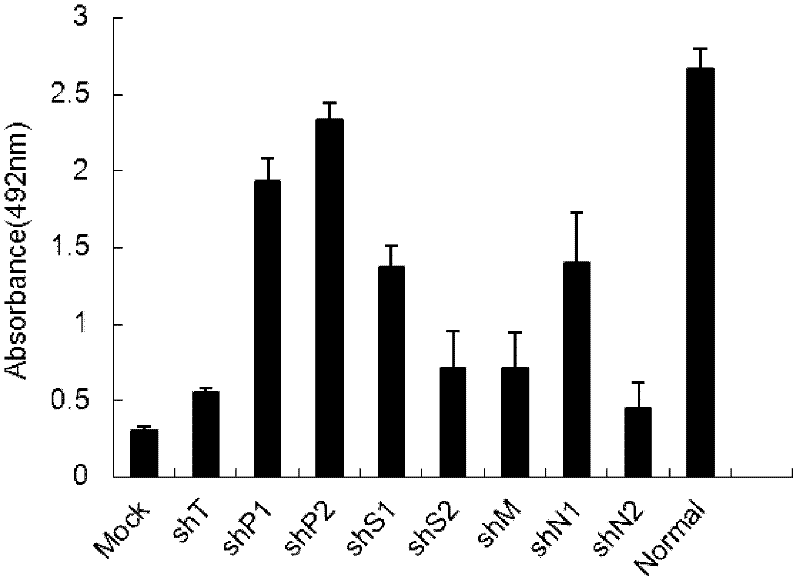
[0036] The shRNA expression plasmid was transfected into ST cells for 28 hours, then the cell culture medium was removed, and 200CCID was used 50 The amount of inoculation TGEV. After 40 hours, analyze cytopathic changes, cytotoxicity (MTS) or extract viral RNA for quantitative RT-PCR detection. .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com