Earthworm protein with HBeAg degrading enzyme activity and application thereof
A technology for degrading enzymes and earthworms, applied in the field of earthworm proteins, can solve the problems of increased drug resistance, unclear other functions of the protein, and the sustained response rate is not as good as that of interferon.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
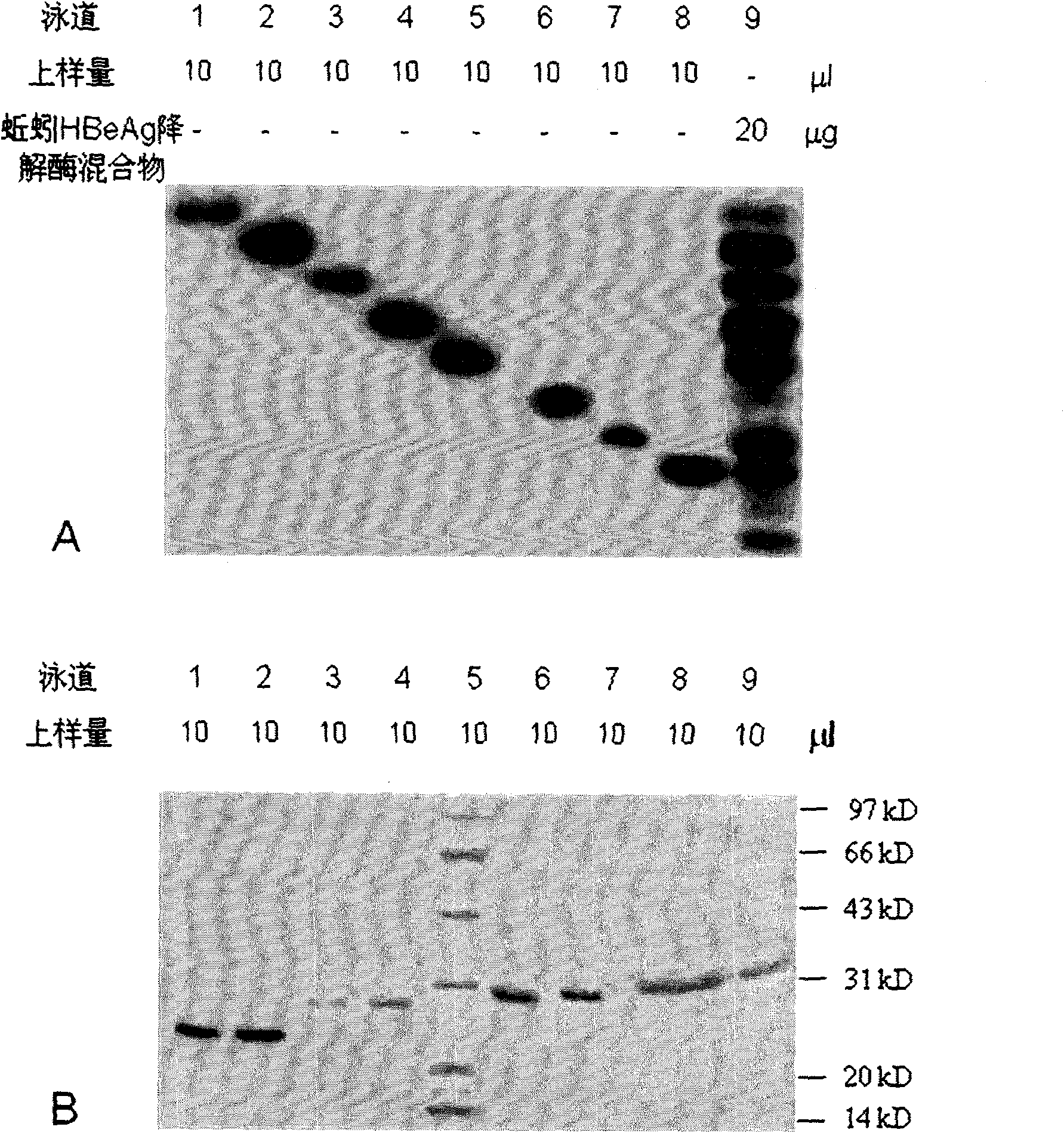
[0069] In one embodiment of the present invention, a kind of preparation method of earthworm protein crude enzyme extract is provided, it comprises the following steps:
[0070] 1) Select 2-month-old, 5-8cm long E. fetida and wash them in distilled water, then soak the earthworms in distilled water at room temperature for 12 hours to make them spit out the dirt in the digestive tract.
[0071] 2) Homogenize 1 kg of earthworms in 0.05M Tris-HCl buffer solution, and centrifuge at 4°C and 8000rpm for 30min. The supernatant was taken and dialyzed against 85% ammonium sulfate equilibrium solution overnight at 4°C.
[0072] 3) Sepharose-4B (10ml) was used as a carrier, activated by carbonyldiimidazole (800mg), and coupled with soybean trypsin inhibitor ligand to prepare an affinity carrier. The prepared affinity carrier was loaded into the column, and 0.1mol / L NaHCO 3 buffer (pH 8.0) to equilibrate the column. The supernatant is adsorbed by the upper affinity chromatography colum...
Embodiment 1
[0083] Embodiment 1 Earthworm protein of the present invention and HBeAg protein co-incubation test
[0084] Experimental methods and results
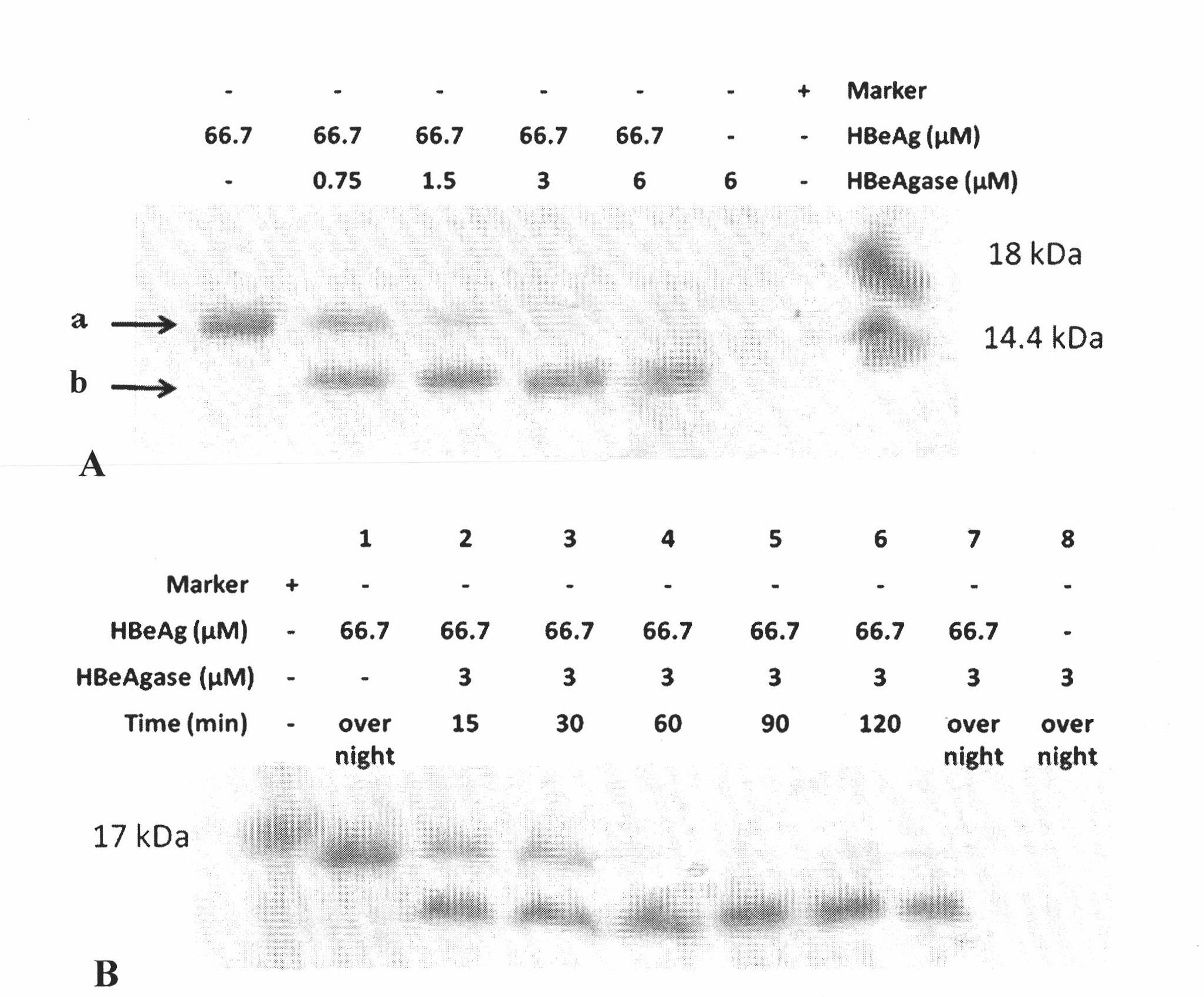
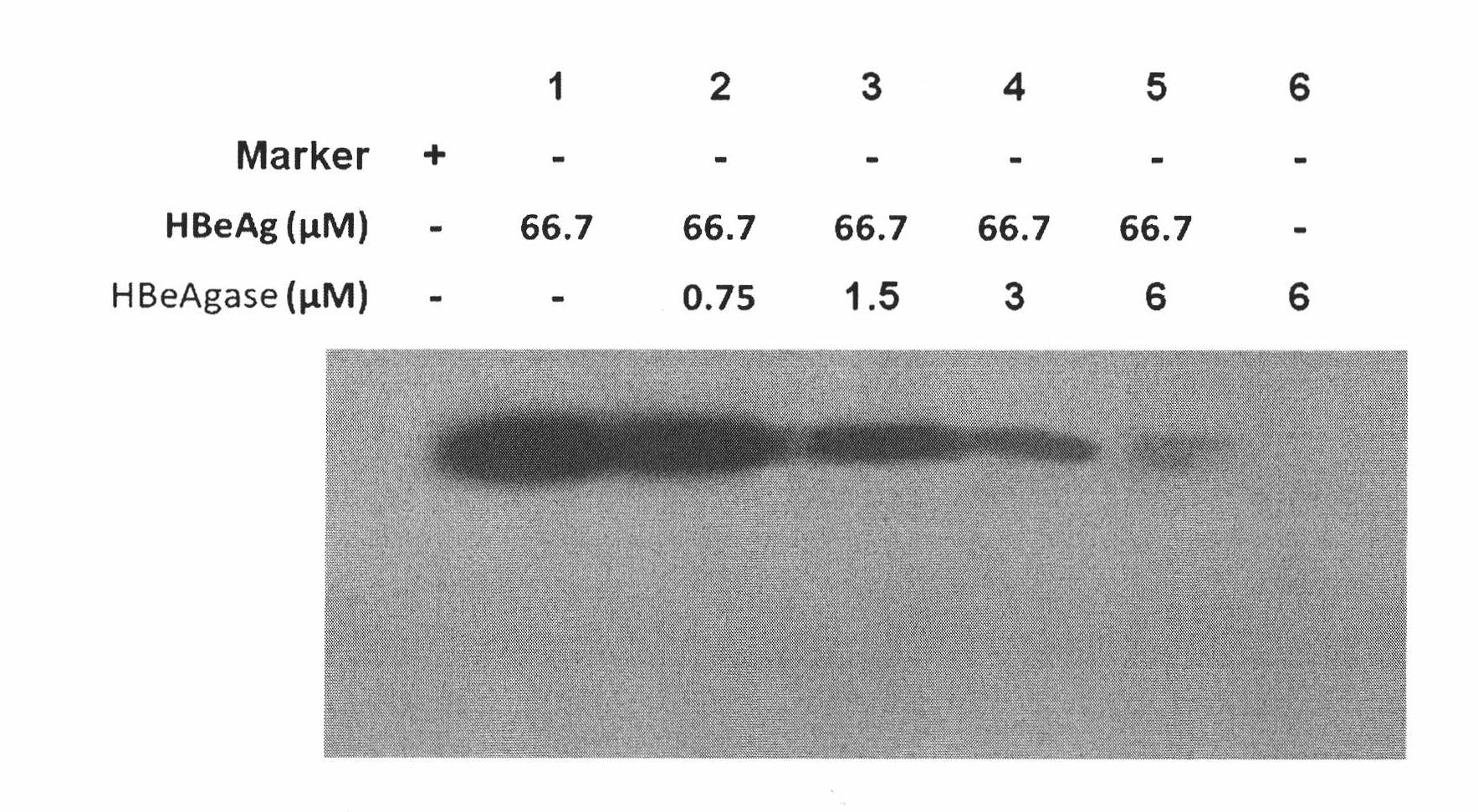
[0085] 1. Get HBeAg (Beijing Huamei Shengke Biotechnology Co., Ltd.) (66.7 μ M), add a certain concentration of earthworm protein (SEQ ID NO.1 protein) (0 ~ 6 μ M) of the present invention and incubate at 37 ° C, respectively at 15, Samples were taken at 30, 45, 60, 90, and 120 minutes, while blank HBeAg and earthworm protein (SEQ ID No.1) of the present invention (incubated at 37° C. for 120 minutes) were used as controls.
[0086] 2. Add the above incubated samples to 5×SDS-PAGE loading buffer, boil for 10 minutes, perform 15% SDS-PAGE, and finally stain with Coomassie brilliant blue.
[0087] 3. SDS-PAGE see figure 1 . Result shows that blank HBeAg hatches 120min protein band without obvious change at 37 ℃, and protein band degrades rapidly in the incubation product of HBeAg and earthworm protein of the present invention (see fi...
Embodiment 2
[0088] Embodiment 2 The analysis of the hydrolysis position of HBeAg proteolysis by earthworm protein of the present invention
[0089] Experimental methods and results
[0090]1. Incubate 66.7 μM HBeAg protein with earthworm protein (SEQ ID No.1) (6 μM) of the present invention at 37° C. for 60 min, and perform 15% SDS-PAGE on the enzymatic hydrolysis product and transfer to PVDF membrane (Millipore, USA).
[0091] 2. The membrane was stained with Coomassie Brilliant Blue, and the main band generated by enzymatic digestion was subjected to N-terminal sequencing (Applied Biosystem Automated Protein Sequencer, Applied Biosystem Inc., USA).
[0092] 3. analyze the effect of the earthworm protein of the present invention on the HBeAg protein by sequencing results (see Table 1) and start from the C-terminus of the HBeAg protein, and analyze the earthworm protein of the present invention on the HBeAg protein according to the molecular weight ratio of the enzymolysis fragment and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com