Method for constructing a nucleic acid library, reagent and kit
A fragment and labeling technology, applied in the field of nucleic acid library establishment, can solve problems such as incompatibility of sequencing primers, and achieve the effect of improving throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1, the construction of the DNA library with terminal labeling
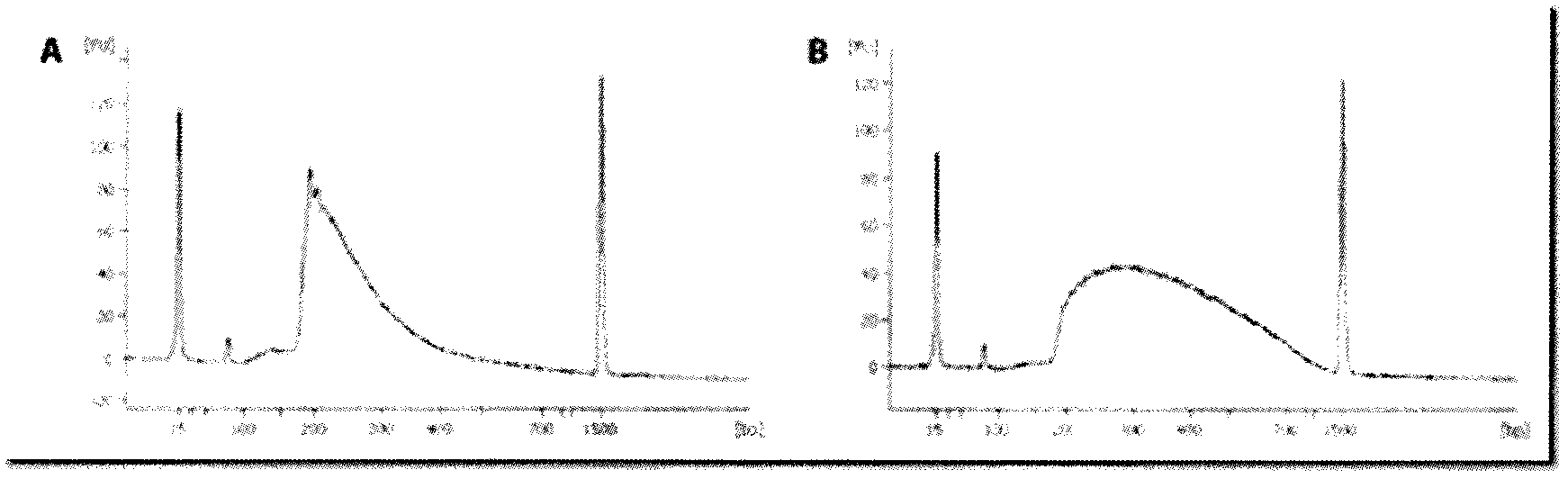
[0022]Use transposase and transposon end sequence (TE) to randomly interrupt the target DNA to make it reach the desired fragment size. Usually, we incubate target DNA, transposase and its specific transposition end sequence in the buffer of transposition reaction. The amount (0.01ng / ul-lug / ul) of target DNA (such as human genomic DNA), transposase (EZ-Tn5 Transposase, EPICENTRE) and its specific transposable end sequence added varies according to different applications. The composition and concentration of the buffer (1×GPSBuffer, 25mM Tris-HCl, 2mM DTT), the temperature and time of incubation also depend on the size of the fragment to be cut. Such as figure 1 Shown: In two different buffer systems, the molecular weight intervals where DNA is broken are different. The reaction can be terminated after adding reaction termination solution (10% sucrose, 66mM EDTA, 20mM TRIS, 0.1% SDS, 0.9% Orang...
Embodiment 2
[0029] Genomic DNA fragments of maize are used as materials, and the target DNA is randomly interrupted by transposase and transposon end sequence (TE) to make it reach the desired fragment size. The buffer system used in the material is (1×GPS Buffer, 25mM Tris-HCl), the samples were processed under different incubation conditions (37°C, 20 minutes), and then the reaction termination solution (10% sucrose, 66mM EDTA, 20mM TRIS , 0.1% SDS, 0.9% Orange G, and 100g / ml proteinase K), the reaction can be terminated after heating at 50°C for 10 minutes. The size of the cleaved DNA fragments can be observed under 1% gel electrophoresis.
[0030] Once the transposition reaction was complete, the transposon end sequence (TE)-tagged DNA fragments were purified using the ZymoDNAClean and Concentrator kit (Zymo Research Corporation, Irvine, CA).
[0031] The purified transposon end sequence (TE)-tagged DNA fragment and a mixture of single-stranded DNA endonuclease (S1nuclease, Promega) ...
Embodiment 3
[0035] Using the genomic DNA of Streptococcus vibrio and the PCR double-stranded DNA fragments circulating in the interrupted Streptococcus DNA as materials, the target DNA was randomly interrupted by transposase and transposon end sequence (TE) to make it achieve our desired fragment size. The buffer system used by the material is ((1×GPS Buffer, 25mM Tris-HCl), the samples were processed under different incubation conditions (25°C, 2 hours), and then the reaction termination solution (10% sucrose, 66mM EDTA, 20mM TRIS , 0.1% SDS, 0.9% Orange G, and 100g / ml proteinase K), the reaction can be terminated after heating at 50°C for 10 minutes. The size of the cleaved DNA fragment can be observed under 1% gel electrophoresis.
[0036] Once the transposition reaction was complete, the transposon end sequence (TE)-tagged DNA fragments were purified using the ZymoDNAClean and Concentrator kit (Zymo Research Corporation, Irvine, CA).
[0037] The purified transposon end sequence (TE)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com