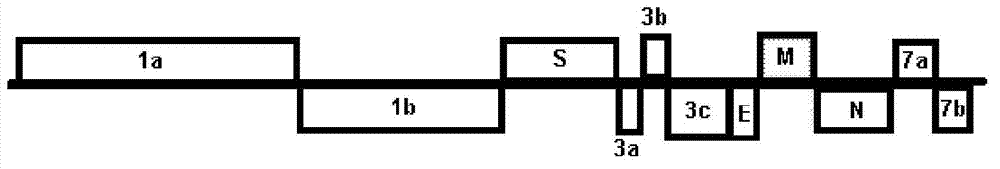
Means and methods for distinguishing FECV and FIPV
A one-part, feline coronavirus technology, applied in biochemical equipment and methods, medical preparations containing active ingredients, pharmaceutical formulations, etc., can solve problems such as inability to distinguish other diseases, inability to qualify, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] In this example, 6 clinical samples (stools) were analyzed. RNA was extracted from clinical samples, RT-PCR was performed on the extracted RNA, and after the first PCR (1st run) and nested PCR (2nd run) it was run by agarose gel electrophoresis (see image 3 ) to analyze the product.
[0073] Materials and methods
[0074] Nested RT-PCR was used to amplify the region of the FCoV spike protein gene containing the targeted mutation. Genomic RNA was extracted from feces of 6 healthy cats using QIAamp Viral RNA Mini Kit and Qiagen RNeasy Mini Kit (Qiagen, Inc.) according to the manufacturer's instructions. cDNA was synthesized using M-MLV reverse transcriptase (RT), followed by polymerase chain reaction (PCR) amplification using Taq DNA polymerase. All enzymes were used according to the manufacturer's instructions (Promega Corp., Madison, WI). Both reactions were primed with specific primers (see Primer Table 1). Primers were designed using the FCoV genome sequences wi...
Embodiment 2
[0078] In this example, fecal or plasma samples from 47 healthy cats, clinical samples from 54 cats with proven FIP, and fecal samples from 14 cats with proven FIP were analyzed.
[0079] Materials and methods
[0080] Extraction of genomic RNA, synthesis, amplification and sequencing of cDNA were carried out according to the materials and methods in Example 1.
[0081] result
[0082] The nucleic acid sequence encoding methionine at amino acid position 1049 was detected in feces or plasma (47 / 47) from all healthy cats ( Figure 4A ). found out later Figure 4A 包含源自证实患有FIP的猫的5条序列(Q093501030_326B_4546.scf、Q093501032_327B_4546.scf、Q093501036_321S_4546.scf、Q093501038_321A_4546.scf和Q093501046_K11_019.ab1),这意味着在来自健康猫的42 / 42源自粪便或 Methionine exists at amino acid position 1049 in FCoV in plasma. 2 / 54 also amplified from cats with proven FIP derived from lesions ( Figure 4B ) RNA and 12 / 14 from feces ( Figure 4C This sequence is observed in the RNA of ). Importantly, it was dem...
Embodiment 3
[0084] We continue to collect samples and cats through Dutch veterinarians and owners. In this example, the following samples were analyzed:
[0085] Fecal samples from 352 healthy cats;
[0086] White blood cell samples from 89 healthy or non-FIP suspicious cats;
[0087] Plasma samples from 89 healthy or non-FIP suspicious cats;
[0088] Serum samples from 56 healthy or non-FIP suspicious cats;
[0089] FIP lesion samples (mesenteric lymph node (LN) and / or kidney and / or spleen and / or omentum and / or lung and / or LN and / or liver and / or ascites) from 97 cats with proven FIP;
[0090] WBC samples from 34 cats with proven FIP;
[0091] Plasma samples from 34 cats with proven FIP; and
[0092] Serum samples from 15 cats with proven FIP.
[0093] Materials and methods
[0094] Extraction of genomic RNA, synthesis, amplification and sequencing of cDNA were carried out according to the materials and methods in Example 1.
[0095] result
[0096] Samples from healthy cats
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com