Microorganism production of high-valve chemical products, and related compositions, methods and systems
A technology of chemical products and microorganisms, applied in the field of metabolic engineering microorganisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
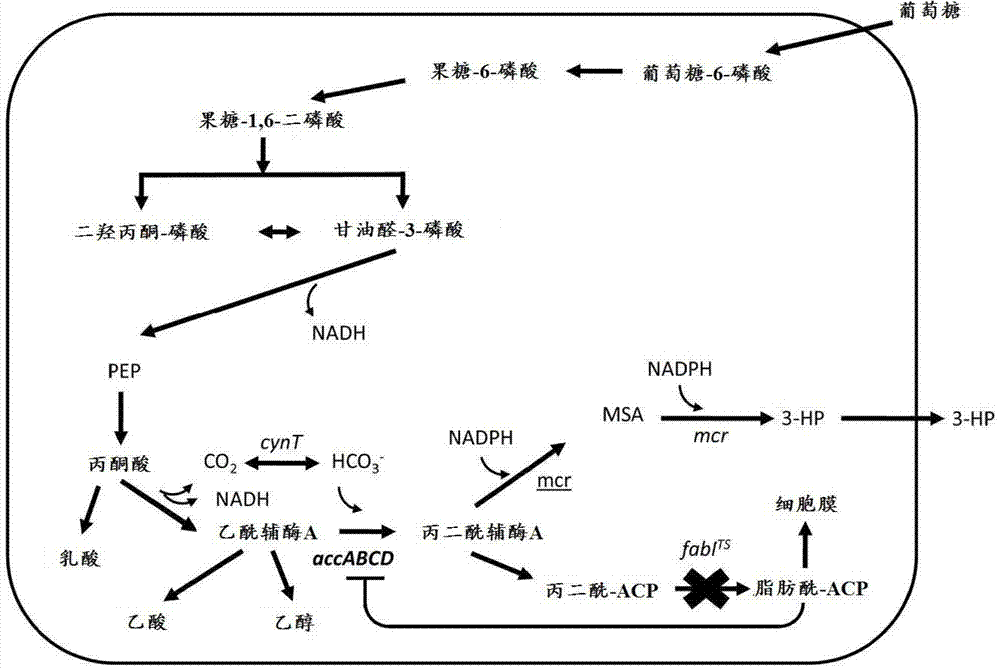
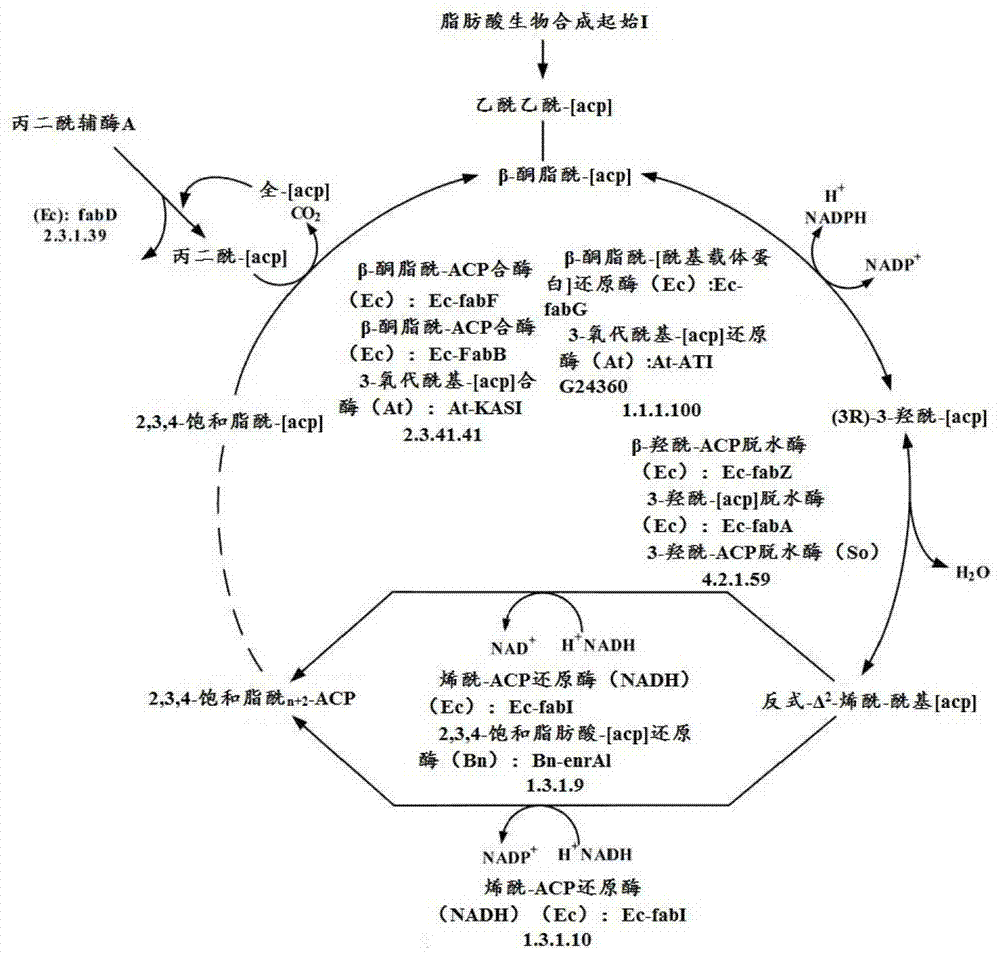
[0464] Therefore, various embodiments of the present invention, such as a method of preparing a chemical, include a conversion step to any such downstream product of 3-HP produced by a microorganism, including but not limited to this article and incorporated references (For regions where the latter is permitted) those chemicals described in the section. For example, one embodiment is to prepare a 3-HP molecule according to the teachings herein, and further convert the 3-HP molecule into polymerized -3-HP (poly-3-HP) or acrylic acid, and for example from the 3-HP molecule The produced acrylic acid is prepared by any of the following: polyacrylic acid (polymerized acrylic acid, in various forms), methyl acrylate, acrylamide, acrylonitrile, propiolactone, ethyl 3-HP, malonic acid, 1,3- Propylene glycol, ethyl acrylate, n-butyl acrylate, hydroxypropyl acrylate, hydroxyethyl acrylate, isobutyl acrylate, 2-ethylhexyl acrylate, and alkyl or aryl groups added thereto and / or added there...
Embodiment 1
[0506] Example 1: Construction of a plasmid expressing malonyl-CoA reductase (mcr)
[0507] According to the service of a commercial DNA gene synthesis provider-DNA2.0 (Menlo Park, CA USA), the nucleotide sequence of the malonyl-CoA reductase gene from Chloroflexus orange was encoded against E. coli Sub-optimization. The gene sequence (SEQ ID NO: 803) introduced an EcoRI restriction site before the start codon and followed by a HindIII restriction site. In addition, a ribosome binding site is set before the start codon. The gene construct was synthesized by DNA2.0 and provided in the pJ206 vector backbone (SEQ ID NO: 804). The plasmid DNA pJ206 containing the synthetic mcr gene was restriction digested with the enzymes EcoRI and HindIII obtained from New England BioLabs (Ipswich, MA USA) according to the manufacturer's instructions. The digestion mixture is separated by agarose gel electrophoresis and the appropriate DNA fragments are recovered as described in the Common Metho...
Embodiment 2
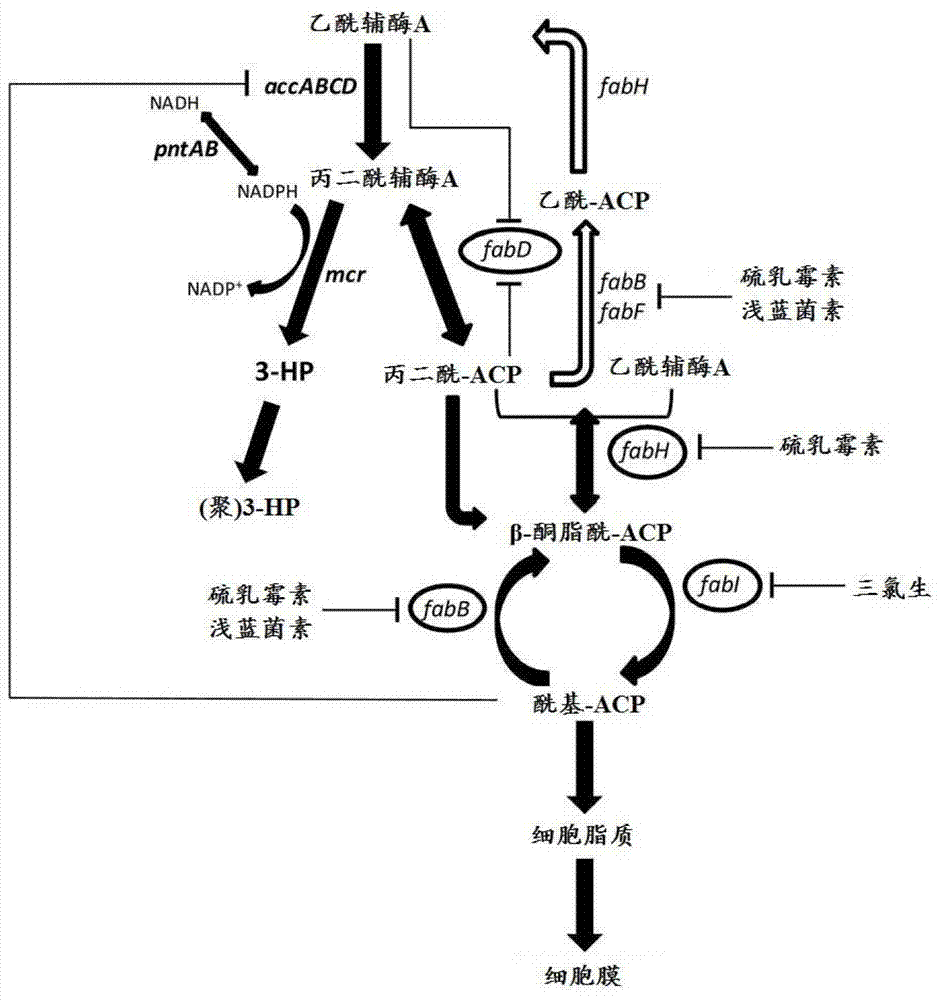
[0511] Example 2: Construction of a plasmid expressing transhydrogenase (pntAB)
[0512] Inducer-independent E. coli promoter derived from the tpiA gene (P tpi A) A fusion with the pyridine nucleotide transhydrogenase gene pntAB (SEQ ID NO: 779 and SEQ ID NO: 781) was created as follows: the tpiA promoter region and pntAB region were amplified from genomic E. coli K12 DNA by polymerase chain reaction . For the pntAB gene, use the pntAB forward primer GGGAACCATGGCAATTGGCATACCAAG (SEQ ID NO: 807, note that all primers disclosed herein are artificial sequences) and the pntAB reverse primer GGGTTACAGAGCTTTCAGGATTGCATCC (SEQ ID NO: 808) to amplify the region, the pntAB The forward primer contains a NcoI site that introduces the starting Met for the pntA protein sequence. Similarly, the PtpiA region was amplified using the forward primer GGGAACGGCGGGGAAAAACAAACGTT (SEQ ID NO: 809) and the reverse primer GGTCCATGGTAATTCTCCACGCTTATAAGC (SEQ ID NO: 810) containing the NcoI restriction s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com