Rapid diagnosis method for plesimonas shigelloides
A Pseudomonas, one-to-one technology, applied in the field of rapid diagnosis of Pseudomonas Shigella, can solve the problem of lower reaction specificity and sensitivity, higher requirements for ratio optimization solution, and higher requirements for primer specificity, etc. problems, to achieve the effects of easy popularization and application, high sensitivity, and improved detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
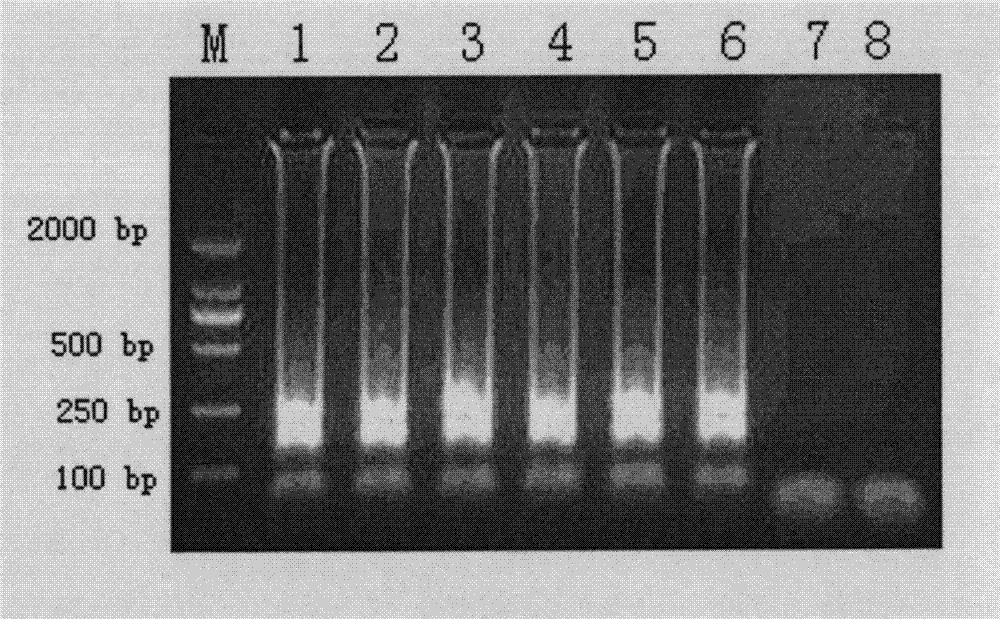
[0044] Embodiment 1: the detection of 4 parts of Shigella-like Plasmomonas
[0045] By comparing the known gene sequences (GeneBank ID number) of P. shigella-like hugA gene in GenBank, we selected a conserved sequence on the hugA gene as the detection target. Using the primer design software PrimerExplorer V4 software ( http: / / primerexplorer.jp ) (Eiken Chemical Co. Ltd., Tokyo, Japan) for LAMP primer design online. Primers were synthesized by Shanghai Sangon Co., Ltd. (see Table 2). The reaction system of LAMP is as follows: the concentration of primers SEQ ID NO 1 and SEQ ID NO 2 is 5pmol, the concentration of primers SEQ ID NO 3 and SEQ ID NO4 is 40pmol, the concentration of primers SEQ ID NO 5 and SEQ ID NO 6 is 20pmol, 10mM Betain, 6mM MgSO 4 , 1 mM dNTP, 2.5 μL of 10×Bst DNA polymerase buffer, 8 U of strand displacement DNA polymerase, 2 μL of template, and add deionized water to 25 μl. The whole reaction was carried out in LA320 real-time turbidimeter (Teramecs, To...
Embodiment 2
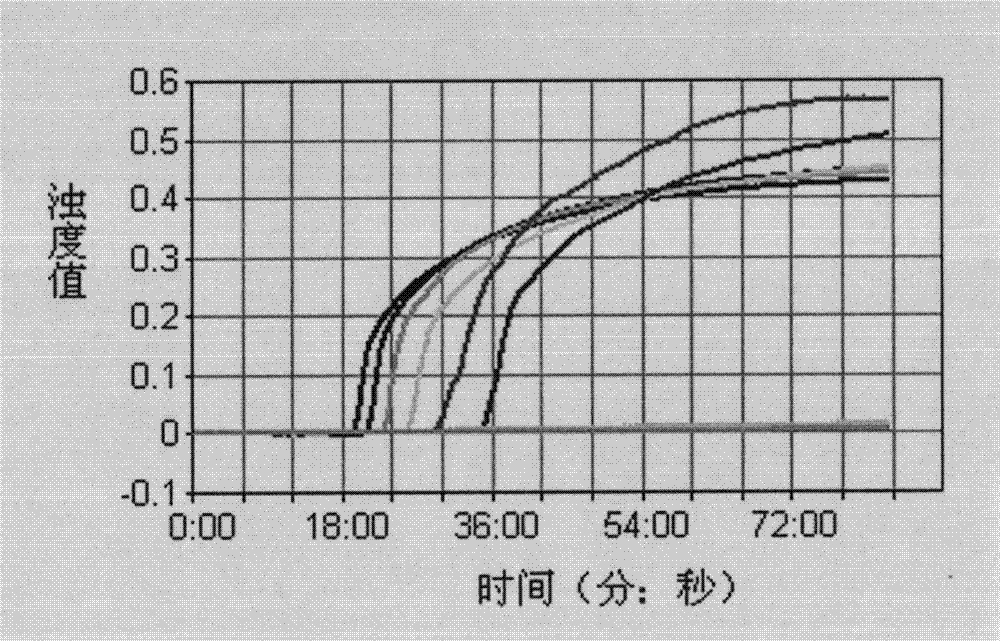
[0047] Embodiment 2: Sensitivity evaluation of LAMP detection:
[0048] 1. Construction and preparation of plasmid standards containing hugA gene
[0049] (1) Use hugA-specific primers for PCR amplification to obtain the target fragment;
[0050] (2) Gel cutting recovery, purification of PCR products;
[0051] (3) connected to the T carrier;
[0052] (4) transform JM109 competent cells;
[0053] (5) Screen positive clones, verify by PCR, and finally confirm by sequencing;
[0054] (6) Extract the plasmid, and convert the concentration of the plasmid sample to the copy number concentration according to the molecular weight of the plasmid: the copy number of the detected gene in each μL sample=concentration (ng / μL)×Avogadro’s constant×10 -9 / (660×recombinant plasmid base number);
[0055] (7) Use Easy Dilution to dilute the above plasmids to 1.0×10 0 Copy / μL~1.0×10 6 copies / μL, a total of 7 concentration gradients.
[0056] 2. Sensitivity of LAMP detection system
[005...
Embodiment 3
[0063] Example 3: Evaluation of the specificity of LAMP detection: the specificity of the LAMP system was evaluated using DNA of common pathogenic bacteria and conditional pathogenic bacteria (see Table 2) as templates.
[0064]The specificity of LAMP detection was evaluated using the DNA of 52 strains of common pathogenic bacteria and conditional pathogenic bacteria (see Table 2). The 20 strains of P. shigella-like bacteria all showed positive amplification within 1 hour, and the 32 strains of non-P. shigella-like bacteria showed no amplification, which indicated that the specificity of the LAMP detection technique was good.
[0065] Bacterial strains used in the experiment of table 1
[0066]
[0067]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com