Biomarker for mass colorectal cancer screening
A biomarker, colorectal cancer technology, applied in biochemical equipment and methods, microbial determination/inspection, application, etc., can solve problems such as research on detection methods without RSPO2 gene methylation, and achieve fast processing and cost. Low, convenient and flexible sample collection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
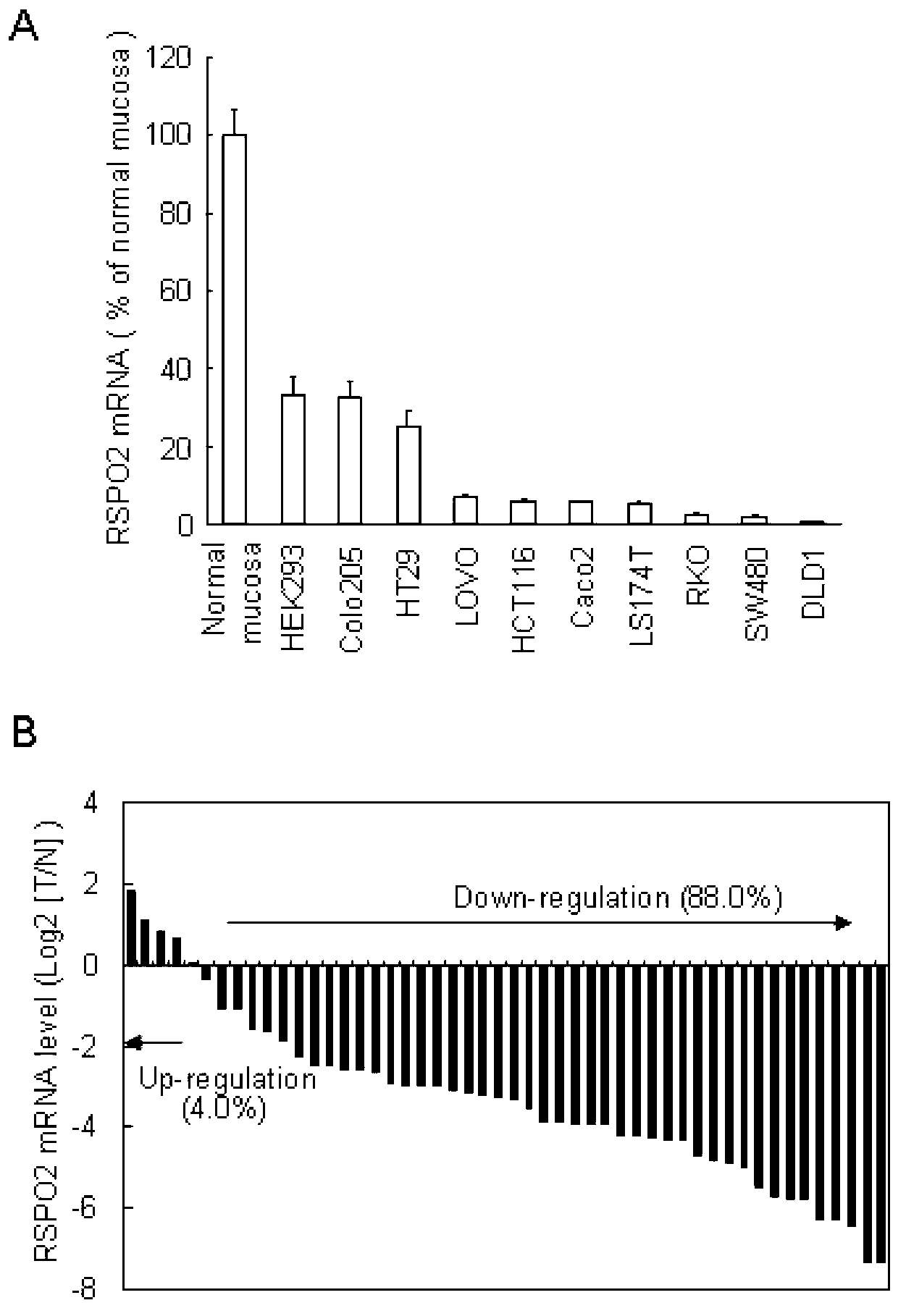
[0037] Down-regulation of RSPO2 expression in colorectal cancer
[0038] Total cellular RNA was extracted by the Trizol method and reverse-transcribed into cDNA by the MLV-reverse transcriptase kit from Invitrogen Company, and all operations were performed according to the method recommended by the kit. Primers RSPO2-F and RSPO2-R for fluorescent quantitative PCR were designed according to the nucleotide sequence of human RSPO2. The primer sequences are as follows:
[0039]Primer RSPO2-F: 5'-ACAATACTGTGTCCAACCAT-3';
[0040] Primer RSPO2-R: 5'-TCCTCTTCTCCTTCGCCTTT-3';
[0041] Using GADPH as an internal reference, the amplification primers are:
[0042] GADPH-F: 5'-ACGGATTTGGTCGTATTGGGC-3';
[0043] GADPH-R: 5'-CTCGCTCCTGGAAGATGGTGAT-3'.
[0044] Using cDNA as a template, the RealMssterMix kit from TIANGEN Company was used for real-time fluorescent quantitative PCR reaction, and the operation was carried out according to the method recommended by the kit.
[0045] Specifi...
Embodiment 2
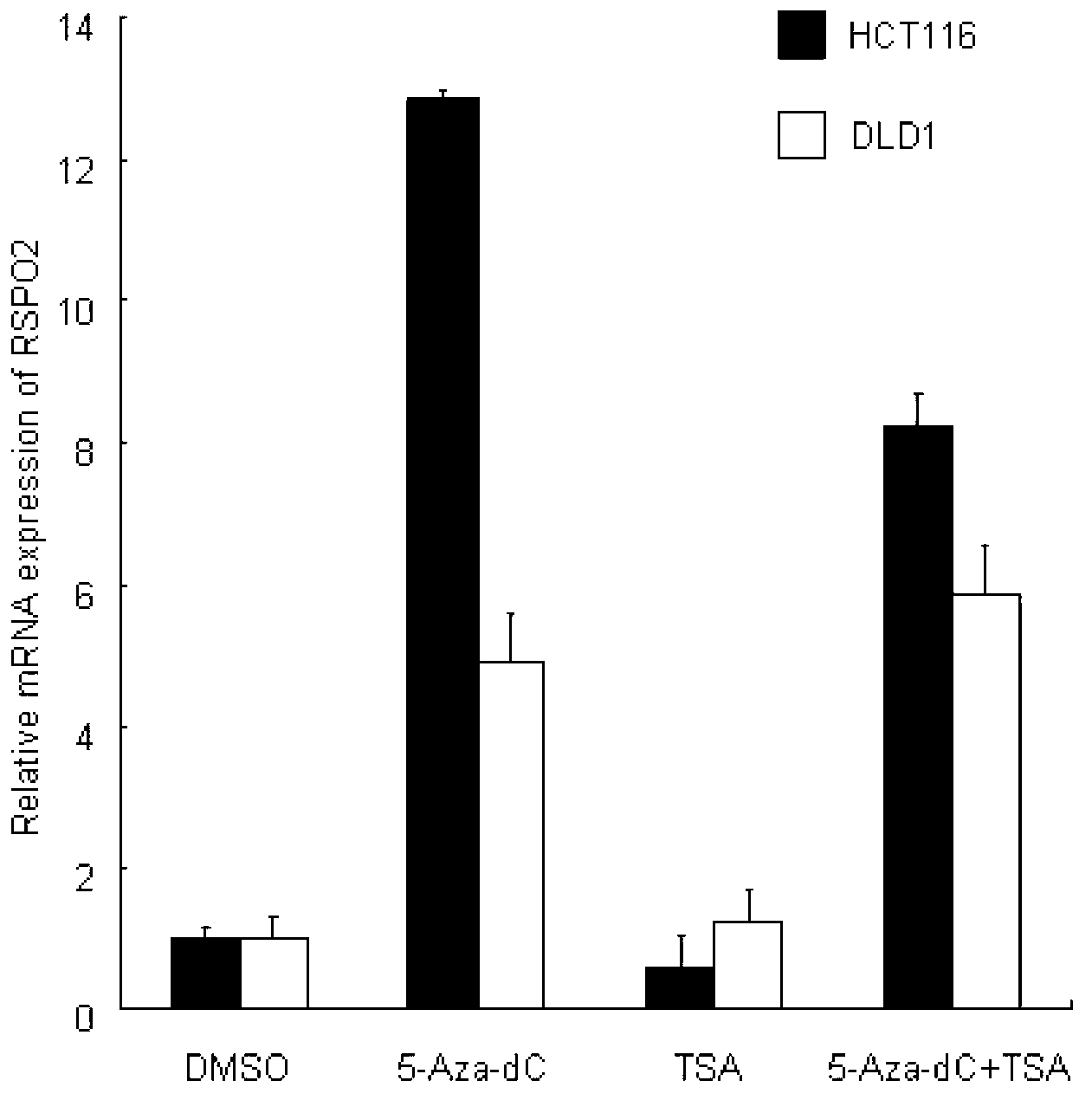
[0052] Identification of RSPO2 methylation as a potential biomarker in colorectal cancer
[0053] (1) The expression level of RSPO2 in intestinal cancer cells was up-regulated after 5-aza-2'-deoxycytidine treatment
[0054] The low expression of RSPO2 in colorectal cancer cells may be caused by epigenetic abnormalities. To distinguish whether it was due to gene methylation or histone modification, we used the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine and the histone deacetylase inhibitor trichostatin, respectively. Intestinal cancer cells were treated with factor A and then the expression of RSPO2 mRNA in the cells was detected.
[0055] Specifically, 1×10 5 Cells / well were seeded in 6-well plates and cultured for 24 h. Experiment with the following groups:
[0056] The control group was the cells without drug addition in the same period;
[0057] test group:
[0058] ①5-Aza-dC group: treated with 2 μM 5-Aza-dC for 96 h,
[0059] ②TSA group: treated with 0....
Embodiment 3
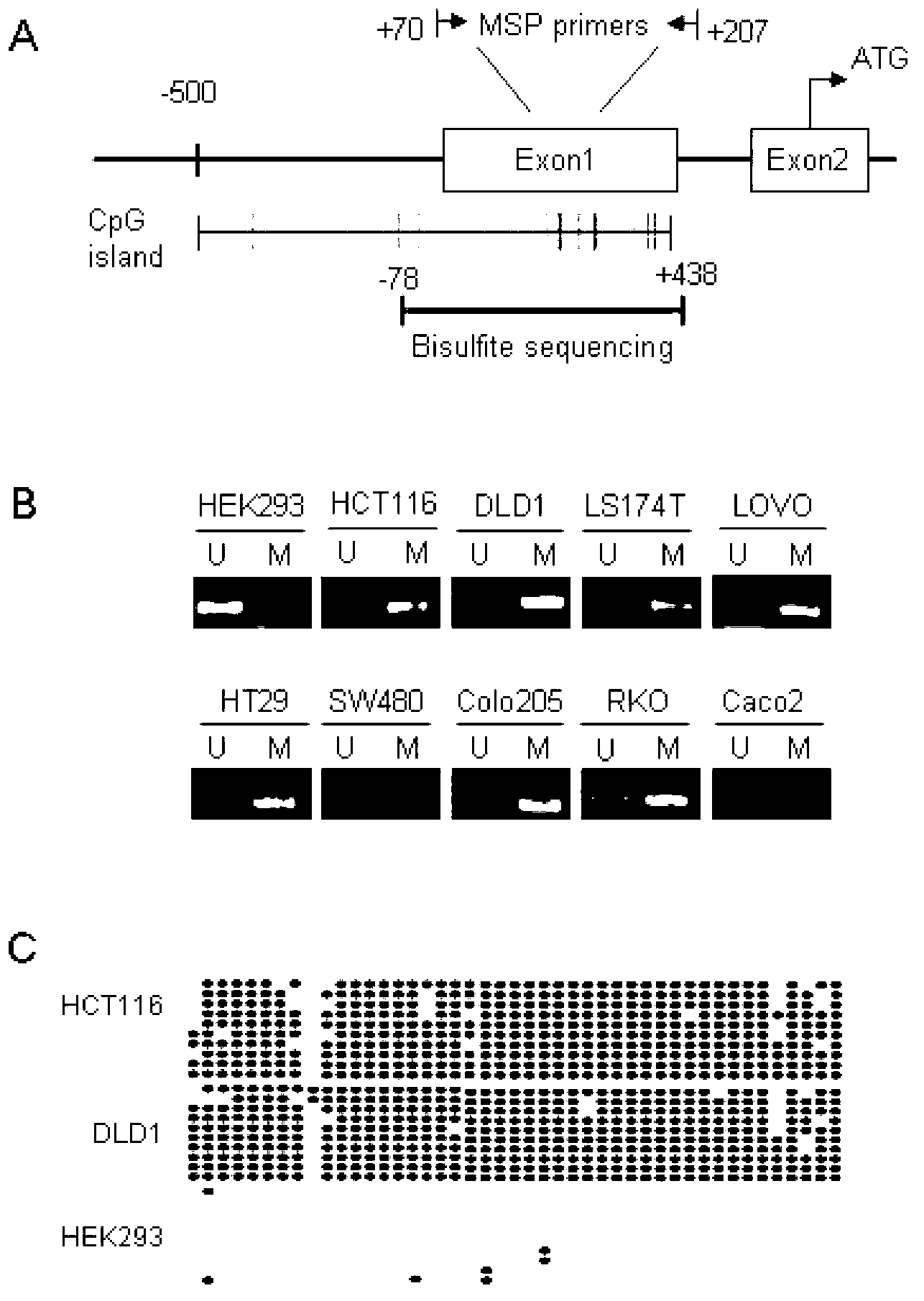
[0083] Application of MSP to detect the methylation status of RSPO2 in colorectal cancer tissues
[0084] Genomic DNA extraction and sodium bisulfite modification were the same as described in Example 2. The MSP method was used to detect the status of RSPO2 methylation in colorectal cancer tissues and normal intestinal mucosal tissues. Specifically, using the modified DNA as a template, using methylated primers, RSPO2-M1 and RSPO2-M2, and unmethylated specific primers, RSPO2-U1 and RSPO2-U2, to perform PCR amplification to amplify the fragment Both were 139 bp in size.
[0085] The PCR reaction system is 20 μl: 10×PCR buffer 2 μl, Taq enzyme 0.5 μl, 10 mM dNTP 2 μl, DNA 60 ng, 10 mM primers 1 μl each, ddH 2 O was added to 20 μl. The PCR reaction conditions were: pre-denaturation at 94°C for 3 min, 37 cycles of 94°C for 30 s, annealing for 45 s, and 72°C for 40 s, and extension at 72°C for 10 min. PCR products were detected by 2% agarose gel electrophoresis.
[0086] MSP m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com