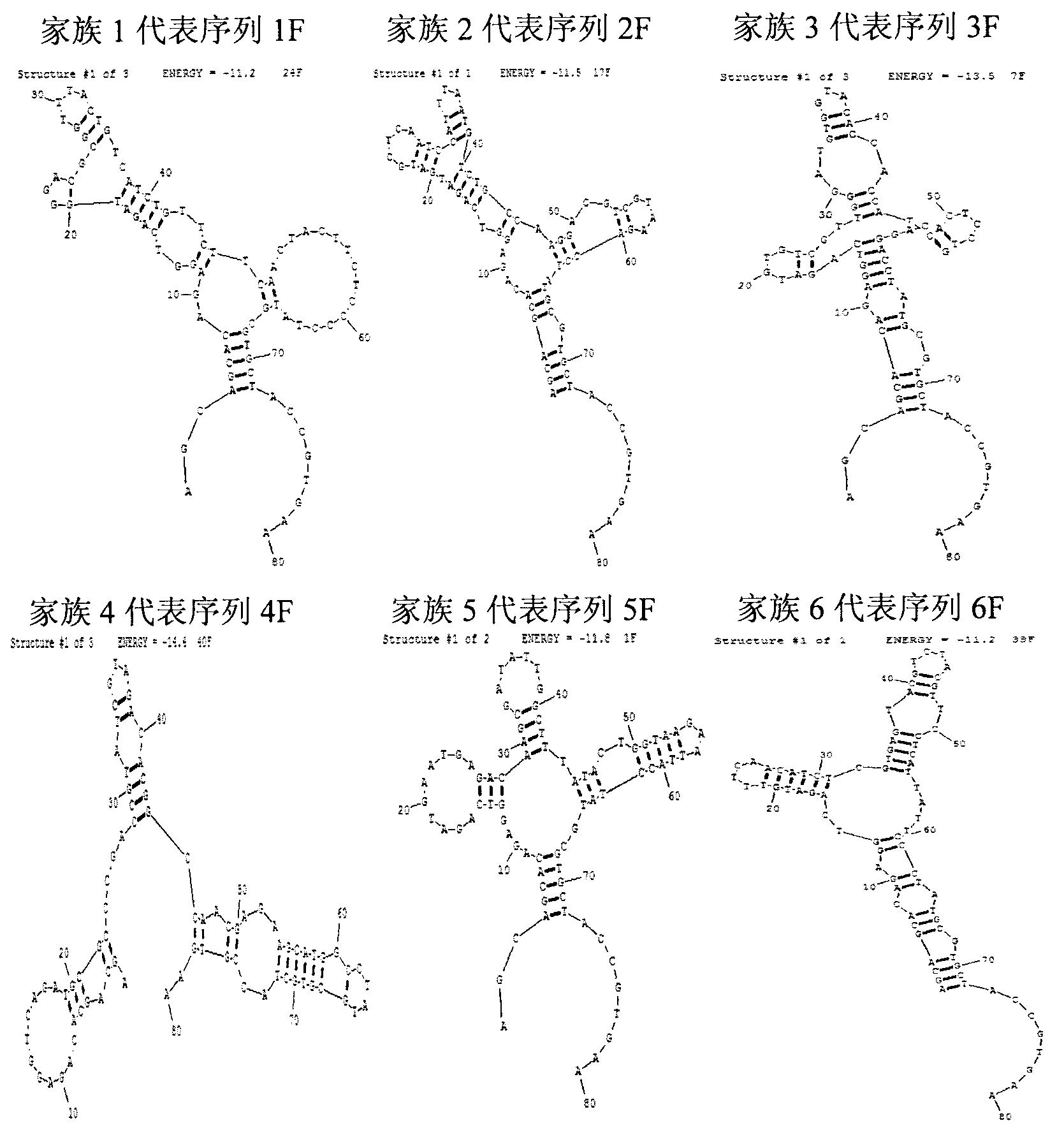
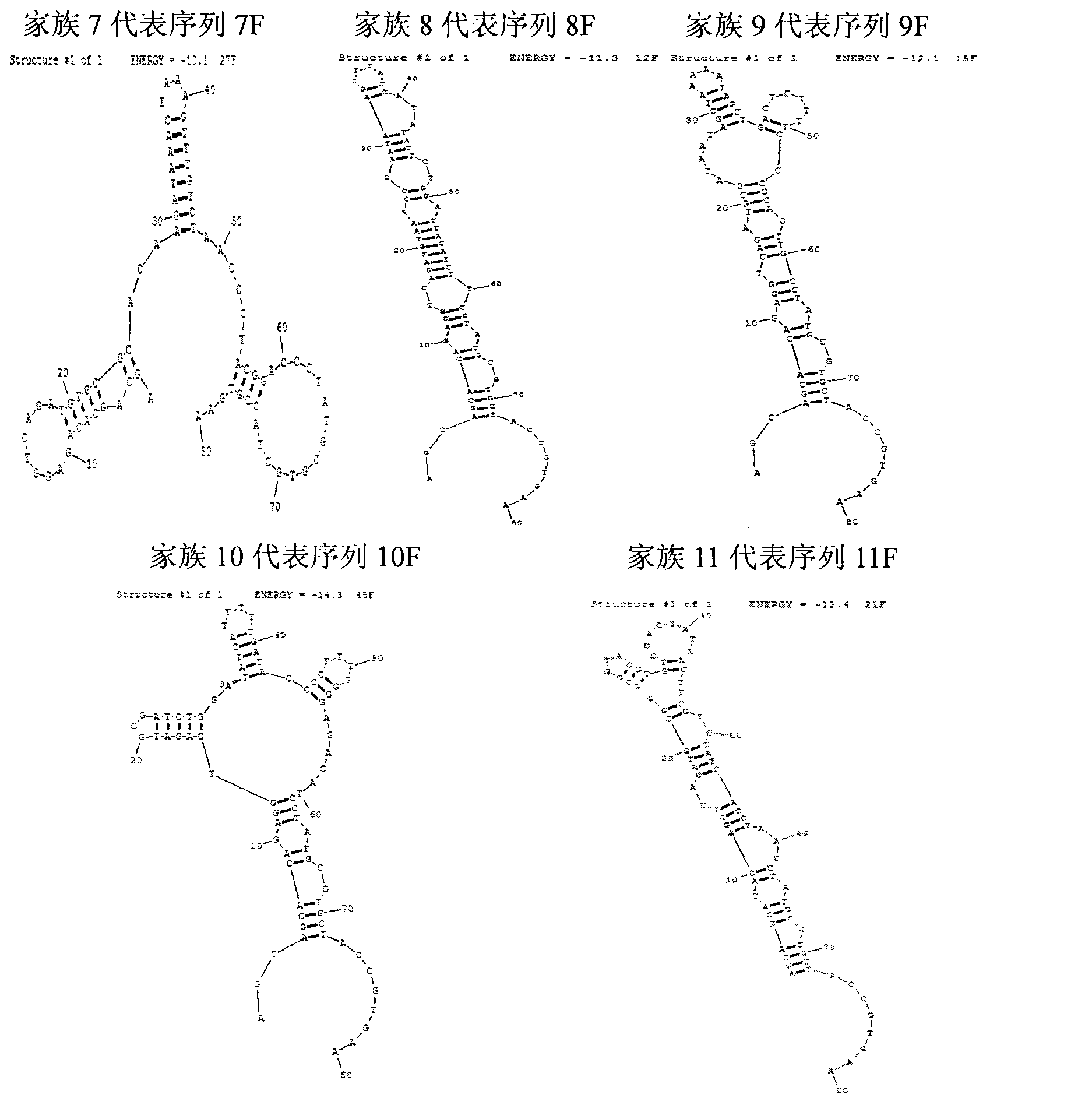
Oligonucleotides aptamer special for distinguishing fumonisin B1
A fumonisin, specific recognition technology, applied in the biological field, can solve the problems of high cost, complicated operation, low sensitivity, etc., and achieve the effects of high purity and efficiency, simple preparation method and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1: Competitive SELEX screening of fumonisin B1 specific binding oligonucleotide aptamers
[0020] 1. In vitro chemical synthesis of initial random single-stranded DNA (ssDNA) library and primers (completed by IDT, USA)
[0021] The sequence is as follows: 5'-AGCAGCACAGAGGTCAGATG(40N)CCTATGCGTGCTACCGTGAA-3'(40N represents 40 random nucleotides);
[0022] Upstream primer: 5′-AGCAGCACAGAGGTCAGATG-3′
[0023] Downstream primer: 5′-TTCACGGTAGCACGCATAGG-3′
[0024] 5′ phosphorylation downstream primer: 5′-P-TTCACGGTAGCACGCATAGG-3′
[0025] The random ssDNA library and primers were made into 100 μM stock solution with TE buffer and stored at -20°C for later use.
[0026] 2. Conditions for PCR amplification and Lambda exonuclease digestion to prepare single-strand sub-libraries
[0027] The synthetic random single-stranded library (ssDNA) was diluted as a PCR template to amplify the phosphorylated double-stranded DNA (dsDNA) product, and the influencing factors of L...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com