Isothermal detection method of RNA (Ribonucleic Acid)
A detection method, isothermal technology, applied in the field of thermally amplified RNA, can solve the problems of easy vibration, instability and weak fluorescence of the chemical structure, and achieve the effect of simple operation, convenient operation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
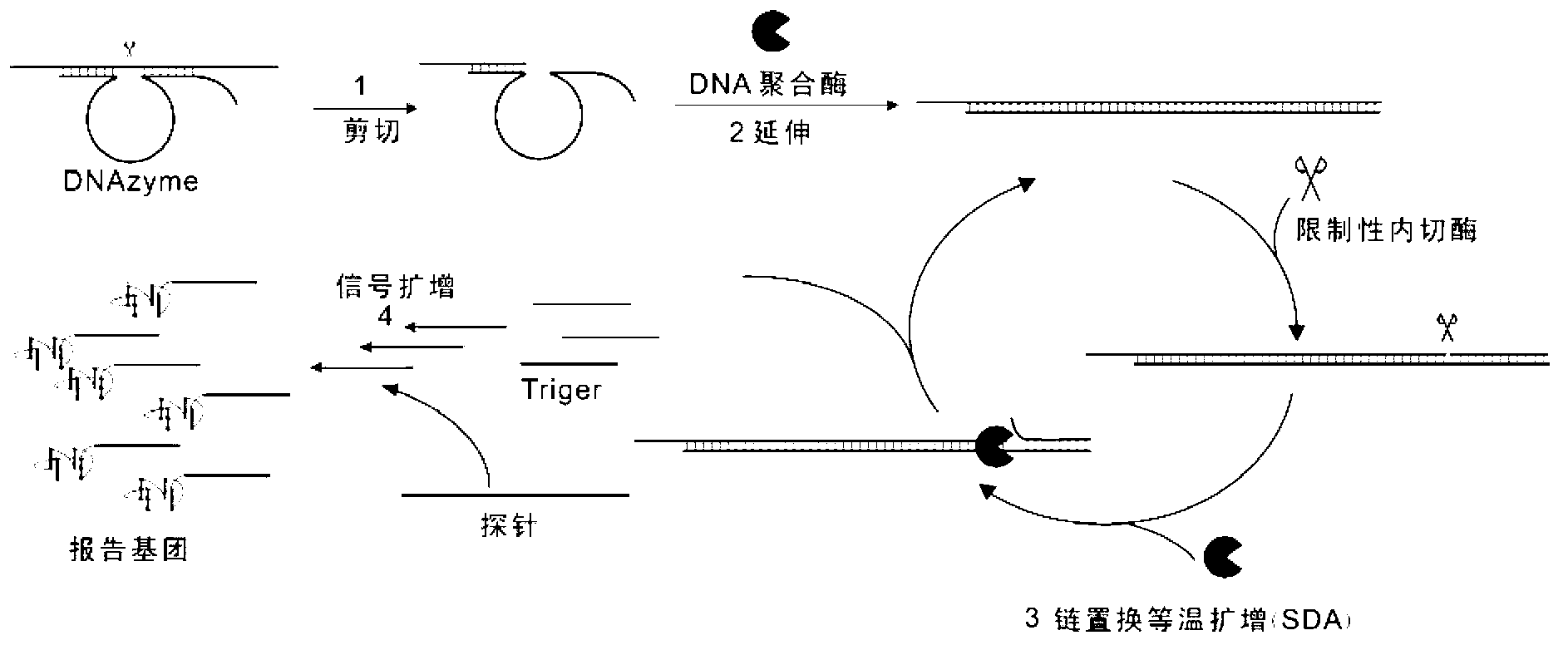
[0043] Example 1: Strand displacement amplification experiment using RNA as primer
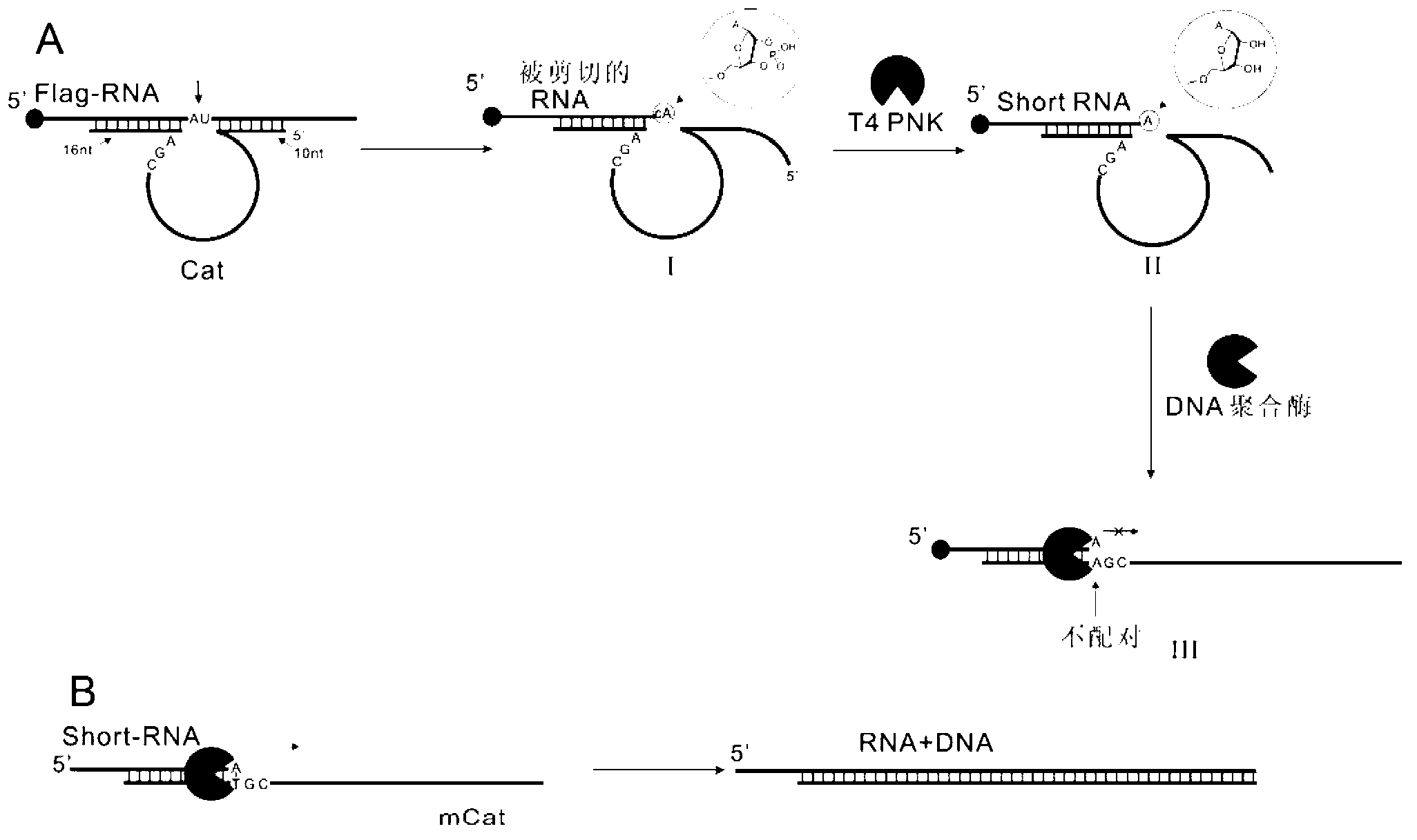
[0044] (1) Extension reaction after RNA is sheared by DNAzyme that shears RNA
[0045] In this experiment, a DNAzyme is firstly used, and the reaction solution 10μL contains 50nm Flag-RNA, 2μM DNAzyme, 10mM Tris-HCl (pH8.5), 100mM KCl, 10mM MgCl 2 , and 0.1mg / ml BSA, 0.5unit / μl PNK and 0.4unit / μl Bsm DNA polymerase, reacted at 37°C for 1h, analyzed with denaturing polyacrylamide gel (PAGE), no extension band appeared. The improved DNAzyme is used here, that is, the bases at the end of the cut RNA are complementary paired with the improved DNAzyme (fCat). After the same reaction conditions as above, extended bands appeared. The function of PNK is to hydrolyze the 2' and 3' cyclic phosphate groups at the cut end of RNA, so that the cut RNA can be used as a primer for extension reaction. as attached figure 2 shown.
[0046] Add a certain concentration of Flag-RNA, 0.5μM fCat, 0.25unit / μl PN...
Embodiment 2
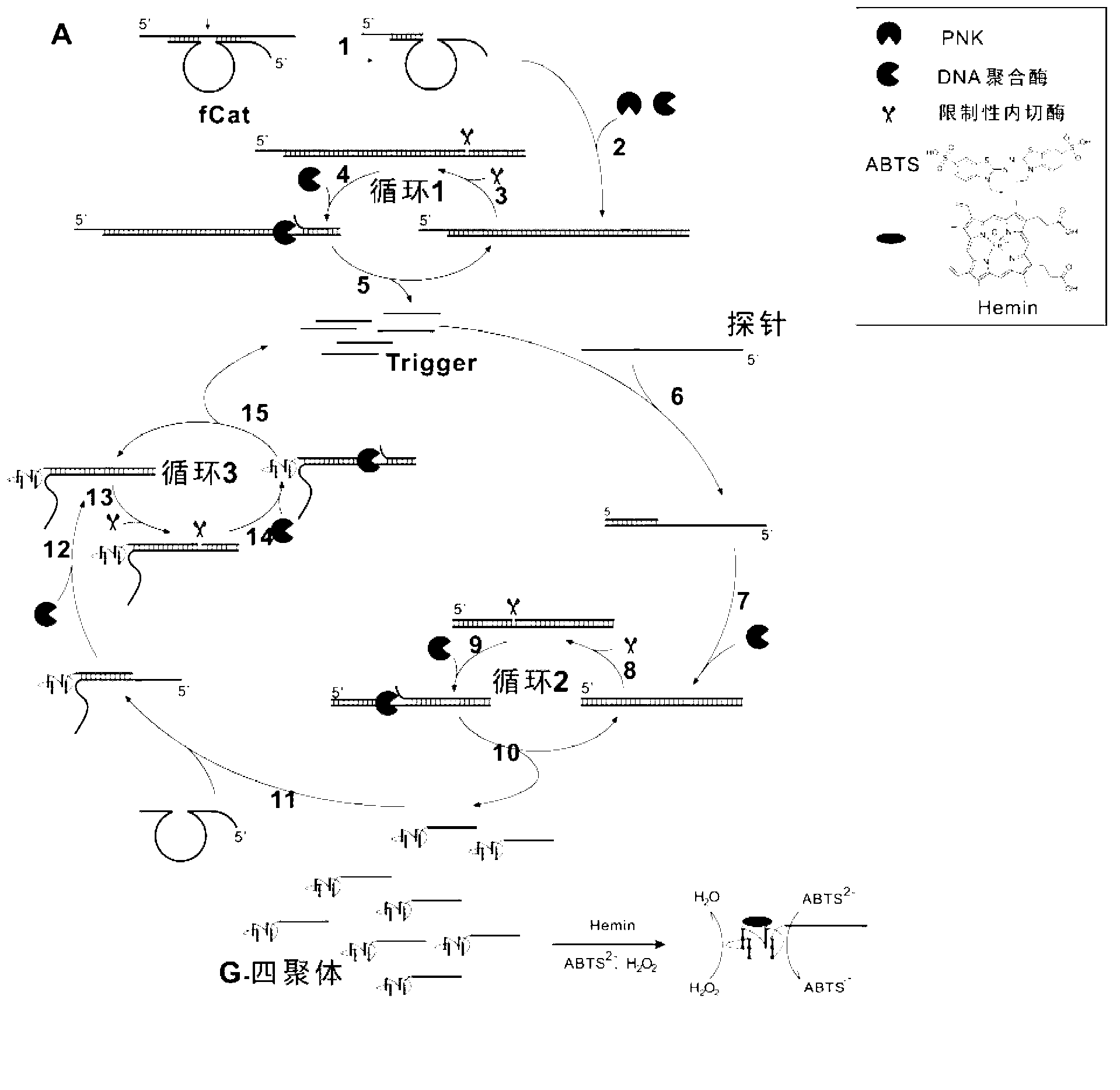
[0067] Example 2: Malachite green combined with Hum21 (a DNAzyme with a G-tetramer structure that can be combined with a dye to generate fluorescence) is developed as a quantitative reporter system experiment
[0068] The quantitative probe Tb of the present invention includes a sequence complementary to trgger from 3' to 5', a restriction endonuclease recognition sequence, a sequence complementary to hum21 that can combine with malachite green to generate fluorescence, and a part of the catalytic center of fCat the same sequence. When the primer is combined with Tb, continuous displacement amplification occurs, thereby producing a large amount of Hum21. Using the property of fluorescence generated by the combination of G-tetramer and malachite green, real-time detection can be achieved with the accumulation of signal. Therefore, it shows that malachite green combined with Hum21 can be developed as a quantitative reporter system.
[0069] Reaction solution 20μL, including 10m...
Embodiment 3
[0072] Example 3 The experiment of using DNAzyme and quantitative probe to quantitatively detect RNA
[0073] For the quantitative detection of RNA, the present invention adopts a one-step method to implement in a reaction container, which further simplifies and speeds up the operation and reduces the risk of contamination at the same time. In the one-step reaction, the improved DNAzyme (fCat) first cuts the RNA, and the function of using PNK is to hydrolyze the 2' and 3' cyclic phosphate groups at the cut end of the RNA, and then the RNA is amplified by continuous displacement. The purpose of detecting RNA is achieved by the quantitative reporter system.
[0074] Reaction solution 20μL, 10mM Tris-HCl, pH7.0, 100mM KCl, 10mM MgCl 2 ,0.1mg / ml BSA, dNTPs (250μM), Nb.Bpu10I (0.17unit / μl), Bsm polymerase (0.17unit / μl), PNK (0.08unit / μl), malachite green (15μmol), 0.25μM fCat, 0.25μM Tb, a certain concentration of Flag-RNA.
[0075] 1 negative control tube: just don't add Flag-R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com