PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) method for identifying seven sea cucumber species
A PCR-RFLP, sea cucumber technology, applied in the field of identifying sea cucumber species, can solve the problems of low throughput, long time, heavy workload, etc., and achieve the effects of good specificity, short typing time, and easy operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
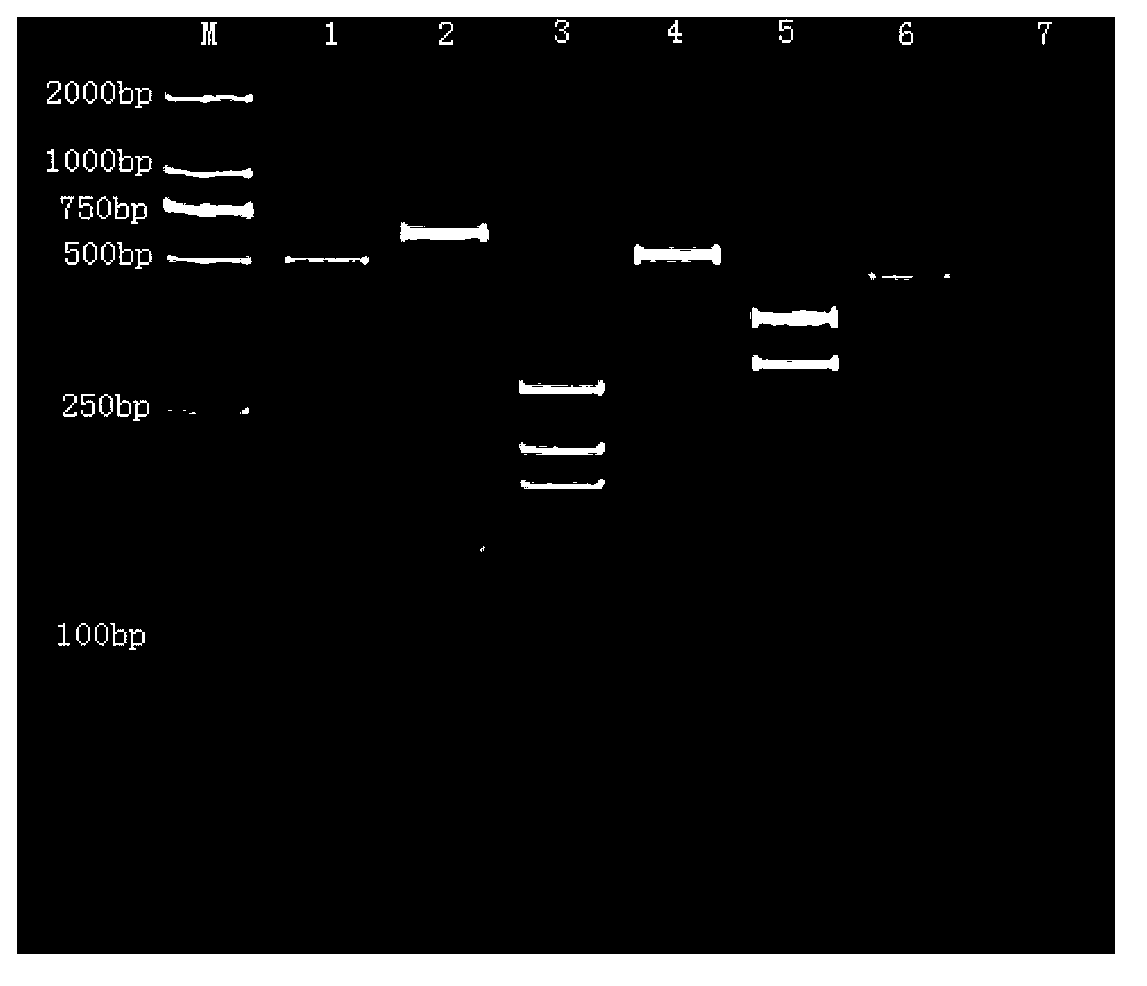
[0036] Identification of known sea cucumber species
[0037] The extracted sea cucumber DNA was amplified by PCR, and the amplified product was subjected to restriction endonuclease digestion experiment. The total PCR-RFLP reaction system was 30 μL, including: 2 μL of 10× enzyme digestion buffer, 10 μL of PCR product, and the corresponding specific Concentration of specific endonuclease: 1FDU / μL0.5μL (for example, the specific endonuclease corresponding to California sea cucumber: SSPⅠ; the specific endonuclease corresponding to North Atlantic sea cucumber: EcoRⅤ; the specific endonuclease corresponding to sea cucumber : SSPⅠ; the specific endonuclease corresponding to plum blossom ginseng: EcoRⅤ; the specific endonuclease corresponding to Haitian melon: ScaⅠ; the specific endonuclease corresponding to American ginseng: PvuⅡ; the specific endonuclease corresponding to boot ginseng: EcoRⅤ, SSPⅠ), the rest were made up with water, 37°C for 30min, and the digested products were d...
Embodiment 2
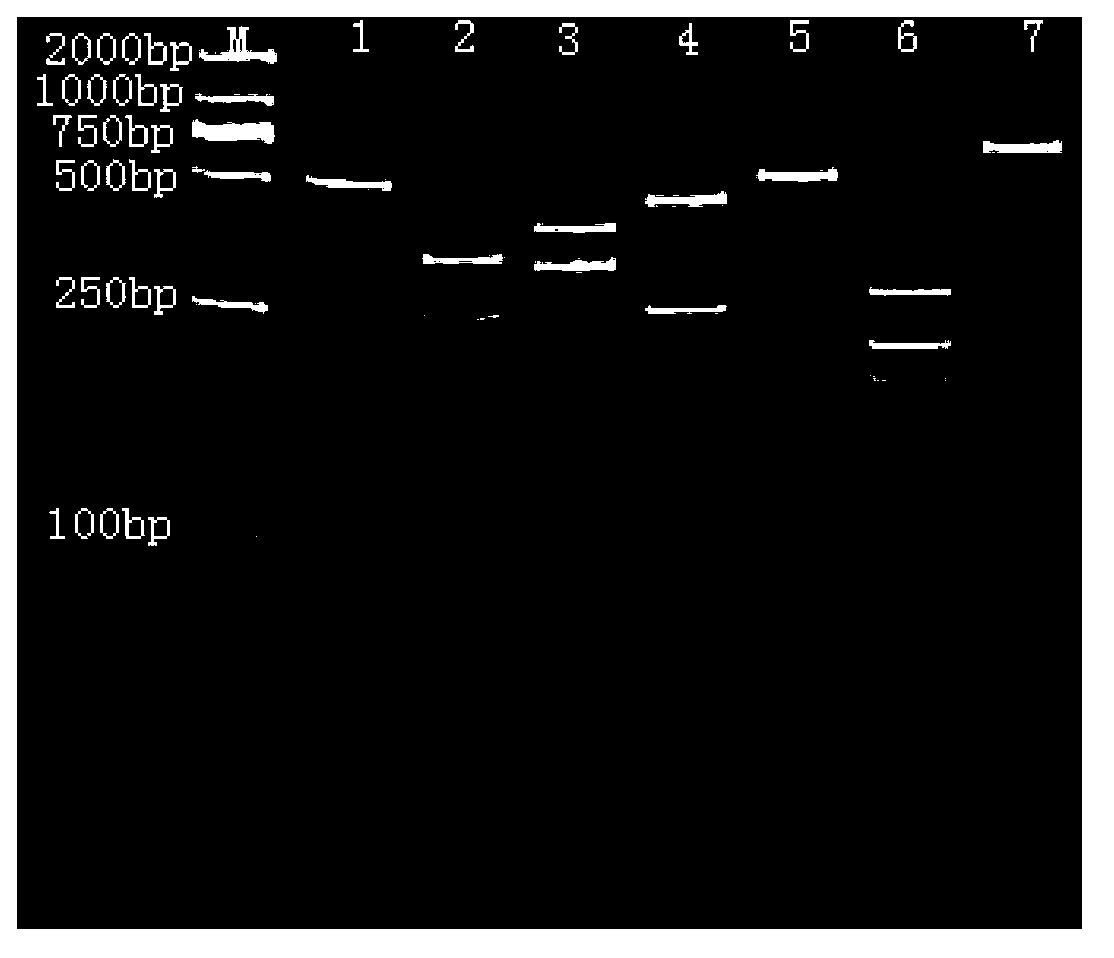
[0040] Identification of Unknown Sea Cucumber Species
[0041] The extracted sea cucumber DNA was amplified by PCR, and the amplified product was subjected to a restriction enzyme digestion experiment. The total reaction system was 30 μL, including: 2 μL of 10× enzyme digestion buffer, 10 μL of PCR product, and the concentration of each of the four enzymes Add 0.5 μL to each 1FDU / μL, 16 μL water, 30 min at 37°C, and detect the digested product by polyacrylamide gel electrophoresis. According to the enzyme digestion results, it was judged that the unknown sample belonged to California japonicus, North Atlantic melon ginseng, imitation japonicus japonicus, plum flower ginseng, Haitian melon, American meat ginseng or boot ginseng. For example, if two bands with a size of 190bp and 498bp are obtained, the sea cucumber to be identified is Apostichopus californica, and the others are similar.
Embodiment 3
[0043] Identification of sea cucumbers with lost external morphological characteristics
[0044] In order to prove that the method can identify sea cucumbers that have lost their external morphological characteristics, the present invention takes sea cucumber enzymatic hydrolyzate as an example to verify. The extracted DNA from the enzymatic solution was amplified by PCR, and the amplified product was subjected to restriction endonuclease digestion experiment. The total reaction system was 30 μL, including: 2 μL of 10× digestion buffer, 10 μL of PCR product, four enzymes each 0.5 μL of each concentration of 1 FDU / μL, 16 μL of double distilled water, 37°C for 30 min, and the digested products were detected by polyacrylamide gel electrophoresis. The result is as image 3 , the electrophoretic pattern of the digested product was consistent with that of the imitated sea cucumber, that is, three bands with sizes of 190bp, 221bp, and 281bp were obtained, indicating that the enzymat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com